1RWO
 
 | | Crystal structure of human caspase-1 in complex with 4-oxo-3-{6-[4-(quinoxalin-2-ylamino)-benzoylamino]-2-thiophen-2-yl-hexanoylamino}-pentanoic acid | | Descriptor: | 4-OXO-3-{6-[4-(QUINOXALIN-2-YLAMINO)-BENZOYLAMINO]-2-THIOPHEN-2-YL-HEXANOYLAMINO}-PENTANOIC ACID, Interleukin-1 beta convertase | | Authors: | Romanowski, M.J, Waal, N.D, Fahr, B.T, O'Brien, T. | | Deposit date: | 2003-12-16 | | Release date: | 2004-12-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural analysis of caspase-1 inhibitors derived from Tethering.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
1SH2
 
 | | Crystal Structure of Norwalk Virus Polymerase (Metal-free, Centered Orthorhombic) | | Descriptor: | RNA Polymerase | | Authors: | Ng, K.K, Pendas-Franco, N, Rojo, J, Boga, J.A, Machin, A, Alonso, J.M, Parra, F. | | Deposit date: | 2004-02-24 | | Release date: | 2004-03-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of norwalk virus polymerase reveals the carboxyl terminus in the active site cleft.
J.Biol.Chem., 279, 2004
|
|
1S19
 
 | | Crystal structure of VDR ligand binding domain complexed to calcipotriol. | | Descriptor: | CALCIPOTRIOL, Vitamin D3 receptor | | Authors: | Tocchini-Valentini, G, Rochel, N, Wurtz, J.M, Moras, D. | | Deposit date: | 2004-01-06 | | Release date: | 2004-04-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the vitamin D nuclear receptor liganded with the vitamin D side chain analogues calcipotriol and seocalcitol, receptor agonists of clinical importance. Insights into a structural basis for the switching of calcipotriol to a receptor antagonist by further side chain modification.
J.Med.Chem., 47, 2004
|
|
2C6C
 
 | | membrane-bound glutamate carboxypeptidase II (GCPII) in complex with GPI-18431 (S)-2-(4-iodobenzylphosphonomethyl)-pentanedioic acid | | Descriptor: | (2S)-2-{[HYDROXY(4-IODOBENZYL)PHOSPHORYL]METHYL}PENTANEDIOIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mesters, J.R, Barinka, C, Li, W, Tsukamoto, T, Majer, P, Slusher, B.S, Konvalinka, J, Hilgenfeld, R. | | Deposit date: | 2005-11-09 | | Release date: | 2006-02-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Glutamate Carboxypeptidase II, a Drug Target in Neuronal Damage and Prostate Cancer.
Embo J., 25, 2006
|
|
1RWP
 
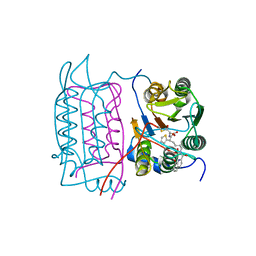 | |
2C6G
 
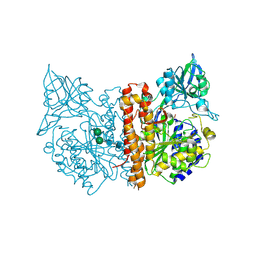 | | Membrane-bound glutamate carboxypeptidase II (GCPII) with bound glutamate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Mesters, J.R, Barinka, C, Li, W, Tsukamoto, T, Majer, P, Slusher, B.S, Konvalinka, J, Hilgenfeld, R. | | Deposit date: | 2005-11-09 | | Release date: | 2006-02-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of glutamate carboxypeptidase II, a drug target in neuronal damage and prostate cancer.
EMBO J., 25, 2006
|
|
4KZ0
 
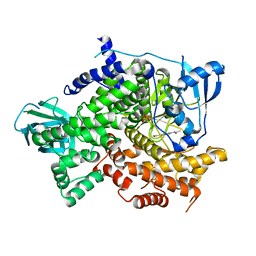 | | Structure of PI3K gamma with Imidazopyridine inhibitors | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION, methyl 2-(acetylamino)-1,3-benzothiazole-6-carboxylate | | Authors: | Knapp, M.S, Elling, R.A. | | Deposit date: | 2013-05-29 | | Release date: | 2013-07-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Structure guided optimization of a fragment hit to imidazopyridine inhibitors of PI3K.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4KZC
 
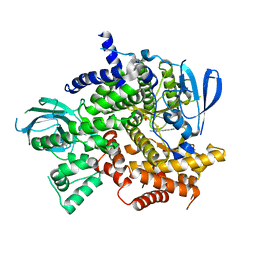 | | Structure of PI3K gamma with Imidazopyridine inhibitors | | Descriptor: | N-{6-[6-amino-5-(trifluoromethyl)pyridin-3-yl]imidazo[1,2-a]pyridin-2-yl}acetamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Knapp, M.S, Elling, E.A. | | Deposit date: | 2013-05-29 | | Release date: | 2013-07-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure guided optimization of a fragment hit to imidazopyridine inhibitors of PI3K.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4M2V
 
 | | Genetically engineered Carbonic Anhydrase IX in complex with Brinzolamide | | Descriptor: | (+)-4-ETHYLAMINO-3,4-DIHYDRO-2-(METHOXY)PROPYL-2H-THIENO[3,2-E]-1,2-THIAZINE-6-SULFONAMIDE-1,1-DIOXIDE, Carbonic anhydrase 2, ZINC ION | | Authors: | Pinard, M.P, Boone, C.D, Rife, B.D, Supuran, C.T, Mckenna, R. | | Deposit date: | 2013-08-05 | | Release date: | 2013-11-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.718 Å) | | Cite: | Structural study of interaction between brinzolamide and dorzolamide inhibition of human carbonic anhydrases.
Bioorg.Med.Chem., 21, 2013
|
|
1S01
 
 | |
1SA8
 
 | |
1SH0
 
 | | Crystal Structure of Norwalk Virus Polymerase (Triclinic) | | Descriptor: | RNA Polymerase | | Authors: | Ng, K.K, Pendas-Franco, N, Rojo, J, Boga, J.A, Machin, A, Alonso, J.M, Parra, F. | | Deposit date: | 2004-02-24 | | Release date: | 2004-03-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal structure of norwalk virus polymerase reveals the carboxyl terminus in the active site cleft.
J.Biol.Chem., 279, 2004
|
|
2BR7
 
 | | Crystal Structure of Acetylcholine-binding Protein (AChBP) from Aplysia californica in complex with HEPES | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, SOLUBLE ACETYLCHOLINE RECEPTOR | | Authors: | Celie, P.H.N, Kasheverov, I.E, Mordvintsev, D.Y, Hogg, R.C, Van Nierop, P, Van Elk, R, Van Rossum-Fikkert, S.E, Zhmak, M.N, Bertrand, D, Tsetlin, V, Sixma, T.K, Smit, A.B. | | Deposit date: | 2005-05-03 | | Release date: | 2005-06-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of Nicotinic Acetylcholine Receptor Homolog Achbp in Complex with an Alpha- Conotoxin Pnia Variant
Nat.Struct.Mol.Biol., 12, 2005
|
|
2BR8
 
 | | Crystal Structure of Acetylcholine-binding Protein (AChBP) from Aplysia californica in complex with an alpha-conotoxin PnIA variant | | Descriptor: | ALPHA-CONOTOXIN PNIA, SOLUBLE ACETYLCHOLINE RECEPTOR, SULFATE ION | | Authors: | Celie, P.H.N, Kasheverov, I.E, Mordvintsev, D.Y, Hogg, R.C, van Nierop, P, van Elk, R, van Rossum-Fikkert, S.E, Zhmak, M.N, Bertrand, D, Tsetlin, V, Sixma, T.K, Smit, A.B. | | Deposit date: | 2005-05-03 | | Release date: | 2005-06-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Nicotinic Acetylcholine Receptor Homolog Achbp in Complex with an Alpha-Conotoxin Pnia Variant
Nat.Struct.Mol.Biol., 12, 2005
|
|
4MI6
 
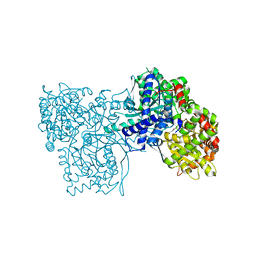 | | Crystal structure of Gpb in complex with SUGAR (N-[4-(5,6,7,8-TETRAHYDRONAPHTHALEN-2-YL)BUTANOYL]-BETA-D-GLUCOPYRANOSYLAMINE) | | Descriptor: | Glycogen phosphorylase, muscle form, N-[4-(5,6,7,8-tetrahydronaphthalen-2-yl)butanoyl]-beta-D-glucopyranosylamine | | Authors: | Kantsadi, L.A, Chatzileontiadou, S.M.D, Leonidas, D.D. | | Deposit date: | 2013-08-30 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure based inhibitor design targeting glycogen phosphorylase b. Virtual screening, synthesis, biochemical and biological assessment of novel N-acyl-beta-d-glucopyranosylamines.
Bioorg.Med.Chem., 22, 2014
|
|
4LWH
 
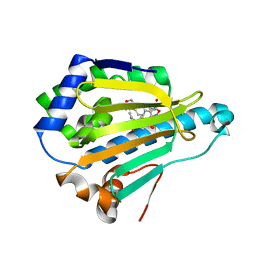 | | Crystal Structure of the human Hsp90-alpha N-domain bound to the hsp90 inhibitor FJ5 | | Descriptor: | Heat shock protein HSP 90-alpha, N-{3-[2,4-dihydroxy-5-(propan-2-yl)phenyl]-4-[4-(morpholin-4-ylmethyl)phenyl]-1,2-oxazol-5-yl}cyclopropanecarboxamide | | Authors: | Li, J, Shi, F, Xiong, B, He, J. | | Deposit date: | 2013-07-27 | | Release date: | 2014-07-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of potent N-(isoxazol-5-yl)amides as HSP90 inhibitors.
Eur.J.Med.Chem., 87, 2014
|
|
4LWG
 
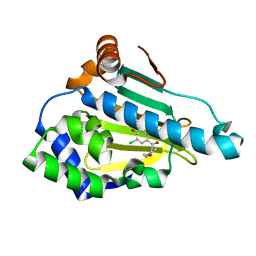 | | Crystal Structure of the human Hsp90-alpha N-domain bound to the hsp90 inhibitor FJ4 | | Descriptor: | 1-(5-chloro-2,4-dihydroxyphenyl)-2-(4-methoxyphenyl)ethanone, Heat shock protein HSP 90-alpha | | Authors: | Li, J, Shi, F, Xiong, B, He, J. | | Deposit date: | 2013-07-27 | | Release date: | 2014-07-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Discovery of potent N-(isoxazol-5-yl)amides as HSP90 inhibitors.
Eur.J.Med.Chem., 87, 2014
|
|
2BIU
 
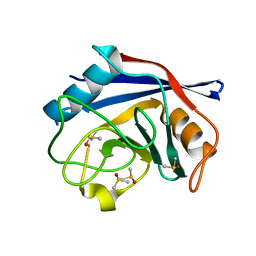 | | Crystal structure of human cyclophilin D at 1.7 A resolution, DMSO complex | | Descriptor: | DIMETHYL SULFOXIDE, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE | | Authors: | Hennig, M, Thoma, R, Stihle, M, Schlatter, D. | | Deposit date: | 2005-01-26 | | Release date: | 2005-01-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystal Engineering Yields Crystals of Cyclophilin D Diffracting to 1.7 A Resolution
Acta Crystallogr.,Sect.D, 61, 2005
|
|
4MBG
 
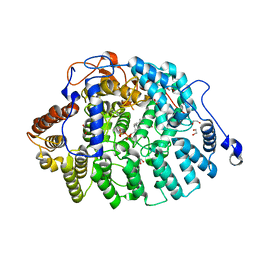 | | Crystal structure of Aspergillus fumigatus protein farnesyltransferase binary complex with farnesyldiphosphate | | Descriptor: | 1,2-ETHANEDIOL, CaaX farnesyltransferase alpha subunit Ram2, CaaX farnesyltransferase beta subunit Ram1, ... | | Authors: | Mabanglo, M.F, Hast, M.A, Beese, L.S. | | Deposit date: | 2013-08-19 | | Release date: | 2014-01-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structures of the fungal pathogen Aspergillus fumigatus protein farnesyltransferase complexed with substrates and inhibitors reveal features for antifungal drug design.
Protein Sci., 23, 2014
|
|
2C67
 
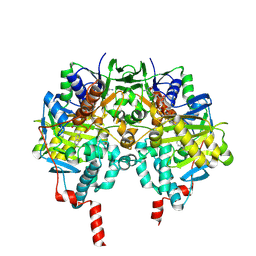 | | MAO inhibition by rasagiline analogues | | Descriptor: | AMINE OXIDASE (FLAVIN-CONTAINING) B, FLAVIN-ADENINE DINUCLEOTIDE, N-METHYL-1(R)-AMINOINDAN | | Authors: | Binda, C, Hubalek, F, Li, M, Herzig, Y, Sterling, J, Edmondson, D.E, Mattevi, A. | | Deposit date: | 2005-11-07 | | Release date: | 2006-01-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Binding of Rasagiline-Related Inhibitors to Human Monoamine Oxidases: A Kinetic and Crystallographic Analysis.
J.Med.Chem., 48, 2005
|
|
2BWX
 
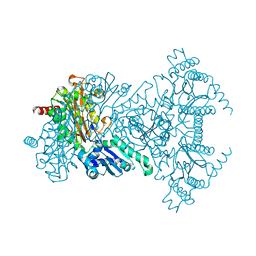 | | His354Ala Escherichia coli Aminopeptidase P | | Descriptor: | AMINOPEPTIDASE P, CHLORIDE ION, MANGANESE (II) ION | | Authors: | Graham, S.C, Guss, J.M. | | Deposit date: | 2005-07-19 | | Release date: | 2006-01-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Kinetic and Crystallographic Analysis of Mutant Escherichia Coli Aminopeptidase P: Insights Into Substrate Recognition and the Mechanism of Catalysis.
Biochemistry, 45, 2006
|
|
2BWV
 
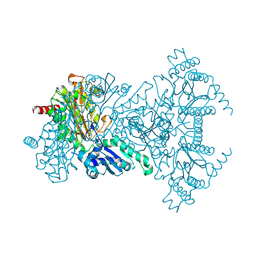 | | His361Ala Escherichia coli Aminopeptidase P | | Descriptor: | AMINOPEPTIDASE P, CHLORIDE ION, MANGANESE (II) ION | | Authors: | Graham, S.C, Guss, J.M. | | Deposit date: | 2005-07-19 | | Release date: | 2006-01-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Kinetic and Crystallographic Analysis of Mutant Escherichia Coli Aminopeptidase P: Insights Into Substrate Recognition and the Mechanism of Catalysis.
Biochemistry, 45, 2006
|
|
2C3I
 
 | | CRYSTAL STRUCTURE OF HUMAN PIM1 IN COMPLEX WITH IMIDAZOPYRIDAZIN I | | Descriptor: | 1-(3-{6-[(CYCLOPROPYLMETHYL)AMINO]IMIDAZO[1,2-B]PYRIDAZIN-3-YL}PHENYL)ETHANONE, PIMTIDE, PROTO-ONCOGENE SERINE THREONINE PROTEIN KINASE PIM1 | | Authors: | Philippakopoulos, P, Knapp, S, Debreczeni, J, Bullock, A, von Delft, F, Sundstrom, M, Arrowsmith, C, Edwards, A, Guo, K, Weigelt, J. | | Deposit date: | 2005-10-07 | | Release date: | 2005-11-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Analysis Identifies Imidazo[1,2- B]Pyridazines as Pim Kinase Inhibitors with in Vitro Antileukemic Activity.
Cancer Res., 67, 2007
|
|
4M2R
 
 | | Human Carbonic Anhydrase II in complex with Brinzolamide | | Descriptor: | (+)-4-ETHYLAMINO-3,4-DIHYDRO-2-(METHOXY)PROPYL-2H-THIENO[3,2-E]-1,2-THIAZINE-6-SULFONAMIDE-1,1-DIOXIDE, Carbonic anhydrase 2, ZINC ION | | Authors: | Pinard, M.P, Boone, C.D, Rife, B.D, Supuran, C.T, Mckenna, R. | | Deposit date: | 2013-08-05 | | Release date: | 2013-11-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.993 Å) | | Cite: | Structural study of interaction between brinzolamide and dorzolamide inhibition of human carbonic anhydrases.
Bioorg.Med.Chem., 21, 2013
|
|
2C5W
 
 | | PENICILLIN-BINDING PROTEIN 1A (PBP-1A) ACYL-ENZYME COMPLEX (CEFOTAXIME) FROM STREPTOCOCCUS PNEUMONIAE | | Descriptor: | 1,2-ETHANEDIOL, CEFOTAXIME, C3' cleaved, ... | | Authors: | Contreras-Martel, C, Job, V, Di Guilmi, A.-M, Vernet, T, Dideberg, O, Dessen, A. | | Deposit date: | 2005-11-02 | | Release date: | 2005-12-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal Structure of Penicillin-Binding Protein 1A (Pbp1A) Reveals a Mutational Hotspot Implicated in Beta-Lactam Resistance in Streptococcus Pneumoniae.
J.Mol.Biol., 355, 2006
|
|
