6Q03
 
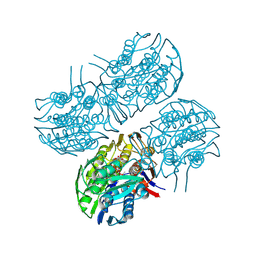 | | Crystal structure of MurA from Clostridium difficile in the presence of UDP-N-acetyl-alpha-D-muramic acid with modified Cys116 (S-[(1S)-1-carboxy-1-(phosphonooxy)ethyl]-L-cysteine) | | Descriptor: | (2R)-2-{[(2R,3R,4R,5S,6R)-3-(acetylamino)-2-{[(S)-{[(R)-{[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}-5-hydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-4-yl]oxy}propanoic acid, 1,2-ETHANEDIOL, UDP-N-acetylglucosamine 1-carboxyvinyltransferase | | Authors: | Dopkins, B.J, Call, C.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2019-08-01 | | Release date: | 2019-11-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of MurA from Clostridium difficile in the presence of UDP-N-acetyl-alpha-D-muramic acid with modified Cys116 (S-[(1S)-1-carboxy-1-(phosphonooxy)ethyl]-L-cysteine)
To Be Published
|
|
6JLR
 
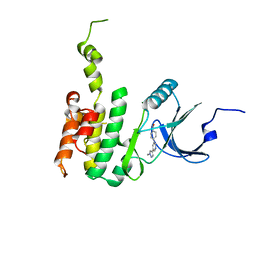 | |
7ZMG
 
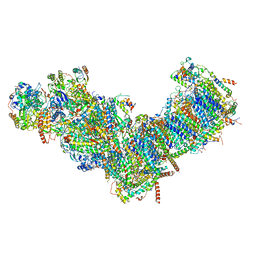 | | CryoEM structure of mitochondrial complex I from Chaetomium thermophilum (state 1) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Laube, E, Kuehlbrandt, W. | | Deposit date: | 2022-04-19 | | Release date: | 2022-11-30 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.44 Å) | | Cite: | Conformational changes in mitochondrial complex I of the thermophilic eukaryote Chaetomium thermophilum.
Sci Adv, 8, 2022
|
|
6CDE
 
 | | Cryo-EM structure at 3.8 A resolution of vaccine-elicited antibody vFP20.01 in complex with HIV-1 Env BG505 DS-SOSIP, and antibodies VRC03 and PGT122 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein 120, ... | | Authors: | Acharya, P, Xu, K, Liu, K, Carragher, B, Potter, C.S, Kwong, P.D. | | Deposit date: | 2018-02-08 | | Release date: | 2018-05-16 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Epitope-based vaccine design yields fusion peptide-directed antibodies that neutralize diverse strains of HIV-1.
Nat. Med., 24, 2018
|
|
2LAV
 
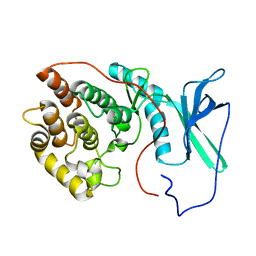 | | NMR solution structure of human Vaccinia-Related Kinase 1 | | Descriptor: | Vaccinia-related kinase 1 | | Authors: | Shin, J, Yoon, H.S. | | Deposit date: | 2011-03-21 | | Release date: | 2011-05-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of Human Vaccinia-related Kinase 1 (VRK1) Reveals the C-terminal Tail Essential for Its Structural Stability and Autocatalytic Activity.
J.Biol.Chem., 286, 2011
|
|
6PHY
 
 | | SpAga D472N structure in complex alpha 1,3 galactobiose | | Descriptor: | 1,2-ETHANEDIOL, Alpha-galactosidase, L(+)-TARTARIC ACID, ... | | Authors: | Pluvinage, B, Boraston, A.B. | | Deposit date: | 2019-06-25 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular analysis of an enigmaticStreptococcus pneumoniaevirulence factor: The raffinose-family oligosaccharide utilization system.
J.Biol.Chem., 294, 2019
|
|
6JOY
 
 | | The X-ray Crystallographic Structure of Branching Enzyme from Rhodothermus obamensis STB05 | | Descriptor: | 1,4-alpha-glucan branching enzyme GlgB | | Authors: | Li, Z.F, Ban, X.F, Jiang, H.M, Wang, Z, Jin, T.C, Li, C.M, Gu, Z.B. | | Deposit date: | 2019-03-25 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.392 Å) | | Cite: | Flexible Loop in Carbohydrate-Binding Module 48 Allosterically Modulates Substrate Binding of the 1,4-alpha-Glucan Branching Enzyme.
J.Agric.Food Chem., 69, 2021
|
|
7T79
 
 | |
6PNO
 
 | |
7TEV
 
 | | Human Ornithine Aminotransferase cocrystallized with its inhibitor, (3S,4R)-3-amino-4-(difluoromethyl)cyclopent-1-ene-1-carboxylate | | Descriptor: | (1S,3R,4S)-3-formyl-4-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]cyclopentane-1-carboxylic acid, Ornithine aminotransferase, mitochondrial | | Authors: | Butrin, A, Zhu, W, Silverman, R, Liu, D. | | Deposit date: | 2022-01-05 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Rational Design, Synthesis, and Mechanism of (3 S ,4 R )-3-Amino-4-(difluoromethyl)cyclopent-1-ene-1-carboxylic Acid: Employing a Second-Deprotonation Strategy for Selectivity of Human Ornithine Aminotransferase over GABA Aminotransferase.
J.Am.Chem.Soc., 144, 2022
|
|
7TFP
 
 | | Human Ornithine Aminotransferase cocrystallized with its inhibitor, (1S,3S)-3-amino-4-(difluoromethylene)cyclopentane-1-carboxylic acid. | | Descriptor: | (1S,3S,4S)-3-amino-4-(fluoromethyl)cyclopentane-1-carboxylic acid, Ornithine aminotransferase, mitochondrial, ... | | Authors: | Butrin, A, Zhu, W, Silverman, R, Liu, D. | | Deposit date: | 2022-01-06 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Rational Design, Synthesis, and Mechanism of (3 S ,4 R )-3-Amino-4-(difluoromethyl)cyclopent-1-ene-1-carboxylic Acid: Employing a Second-Deprotonation Strategy for Selectivity of Human Ornithine Aminotransferase over GABA Aminotransferase.
J.Am.Chem.Soc., 144, 2022
|
|
8HVP
 
 | | STRUCTURE AT 2.5-ANGSTROMS RESOLUTION OF CHEMICALLY SYNTHESIZED HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 PROTEASE COMPLEXED WITH A HYDROXYETHYLENE*-BASED INHIBITOR | | Descriptor: | HIV-1 PROTEASE, INHIBITOR VAL-SER-GLN-ASN-LEU-PSI(CH(OH)-CH2)-VAL-ILE-VAL (U-85548E) | | Authors: | Jaskolski, M, Miller, M, Tomasselli, A.G, Sawyer, T.K, Staples, D.G, Heinrikson, R.L, Schneider, J, Kent, S.B.H, Wlodawer, A. | | Deposit date: | 1990-10-26 | | Release date: | 1993-10-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure at 2.5-A resolution of chemically synthesized human immunodeficiency virus type 1 protease complexed with a hydroxyethylene-based inhibitor.
Biochemistry, 30, 1991
|
|
6Q8Z
 
 | | Structure of human galactokinase 1 bound with N-(Cyclobutylmethyl)-1,5-dimethyl-1H-pyrazole-4-carboxamide | | Descriptor: | 2-(1,3-benzoxazol-2-ylamino)spiro[1,6,7,8-tetrahydroquinazoline-4,1'-cyclohexane]-5-one, Galactokinase, beta-D-galactopyranose, ... | | Authors: | Mackinnon, S.R, Bezerra, G.A, Zhang, M, Foster, W, Krojer, T, Brandao-Neto, J, Douangamath, A, Arrowsmith, C, Edwards, A, Bountra, C, Brennan, P, Lai, K, Yue, W.W. | | Deposit date: | 2018-12-16 | | Release date: | 2019-01-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of human galactokinase 1 bound with N-(Cyclobutylmethyl)-1,5-dimethyl-1H-pyrazole-4-carboxamide
To Be Published
|
|
4Y0H
 
 | | Gamma-aminobutyric acid aminotransferase inactivated by (1S,3S)-3-amino-4-difluoromethylenyl-1-cyclopentanoic acid (CPP-115) | | Descriptor: | 4-aminobutyrate aminotransferase, mitochondrial, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Rui, W, Ruslan, S, Hyunbeom, L, Emma, H.D, Jose, I.J, Neil, K, Richard, B.S, Dali, L. | | Deposit date: | 2015-02-06 | | Release date: | 2015-03-11 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Mechanism of inactivation of gamma-aminobutyric acid aminotransferase by (1S,3S)-3-amino-4-difluoromethylenyl-1-cyclopentanoic acid (CPP-115)
J. Am. Chem. Soc., 2015
|
|
4XFZ
 
 | | Structure of the native full-length HIV-1 capsid protein in complex with PF-3450074 (PF74) | | Descriptor: | CHLORIDE ION, HIV-1 capsid protein, IODIDE ION, ... | | Authors: | Gres, A.T, Kirby, K.A, Sarafianos, S.G. | | Deposit date: | 2014-12-29 | | Release date: | 2015-06-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | STRUCTURAL VIROLOGY. X-ray crystal structures of native HIV-1 capsid protein reveal conformational variability.
Science, 349, 2015
|
|
8GPD
 
 | | Crystal structure of NDM-1 at pH5.5 (Succinate) in complex with hydrolyzed penicillin V | | Descriptor: | (2R,4S)-5,5-dimethyl-2-[(1R)-2-oxidanyl-2-oxidanylidene-1-(2-phenoxyethanoylamino)ethyl]-1,3-thiazolidine-4-carboxylic acid, Metallo beta lactamase NDM-1, POTASSIUM ION, ... | | Authors: | Shi, X, Dai, Y, Zhang, Q, Liu, W. | | Deposit date: | 2022-08-26 | | Release date: | 2023-08-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Interplay between the beta-lactam side chain and an active-site mobile loop of NDM-1 in penicillin hydrolysis as a potential target for mechanism-based inhibitor design.
Int.J.Biol.Macromol., 262, 2024
|
|
8GPC
 
 | | Crystal structure of NDM-1 at pH5.5 (Succinate) in complex with hydrolyzed ampicillin | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Metallo beta lactamase NDM-1, SODIUM ION, ... | | Authors: | Shi, X, Dai, Y, Zhang, Q, Liu, W. | | Deposit date: | 2022-08-26 | | Release date: | 2023-08-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Interplay between the beta-lactam side chain and an active-site mobile loop of NDM-1 in penicillin hydrolysis as a potential target for mechanism-based inhibitor design.
Int.J.Biol.Macromol., 262, 2024
|
|
8GPE
 
 | | Crystal structure of NDM-1 at pH5.5 (Succinate) in complex with hydrolyzed penicillin G | | Descriptor: | (2R,4S)-2-{(R)-carboxy[(phenylacetyl)amino]methyl}-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Metallo beta lactamase NDM-1, POTASSIUM ION, ... | | Authors: | Shi, X, Dai, Y, Zhang, Q, Liu, W. | | Deposit date: | 2022-08-26 | | Release date: | 2023-08-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Interplay between the beta-lactam side chain and an active-site mobile loop of NDM-1 in penicillin hydrolysis as a potential target for mechanism-based inhibitor design.
Int.J.Biol.Macromol., 262, 2024
|
|
4XFX
 
 | |
7TD0
 
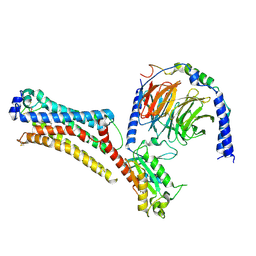 | | Lysophosphatidic acid receptor 1-Gi complex bound to LPA | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, S, Paknejad, N, Zhu, L, Kihara, Y, Ray, D, Chun, J, Liu, W, Hite, R.K, Huang, X.Y. | | Deposit date: | 2021-12-30 | | Release date: | 2022-02-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Differential activation mechanisms of lipid GPCRs by lysophosphatidic acid and sphingosine 1-phosphate.
Nat Commun, 13, 2022
|
|
7TD2
 
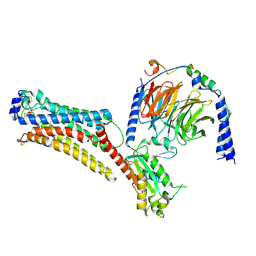 | | Lysophosphatidic acid receptor 1-Gi complex bound to LPA, state a | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, S, Paknejad, N, Zhu, L, Kihara, Y, Ray, D, Chun, J, Liu, W, Hite, R.K, Huang, X.Y. | | Deposit date: | 2021-12-30 | | Release date: | 2022-02-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Differential activation mechanisms of lipid GPCRs by lysophosphatidic acid and sphingosine 1-phosphate.
Nat Commun, 13, 2022
|
|
4XS2
 
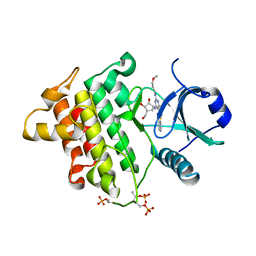 | | Irak4-inhibitor co-structure | | Descriptor: | (1R,2S,3R,5R)-3-({5-(1,3-benzothiazol-2-yl)-6-chloro-2-[(3-methoxypropyl)amino]pyrimidin-4-yl}amino)-5-(hydroxymethyl)cyclopentane-1,2-diol, Interleukin-1 receptor-associated kinase 4 | | Authors: | Fischmann, T.O. | | Deposit date: | 2015-01-21 | | Release date: | 2015-05-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Discovery and hit-to-lead optimization of 2,6-diaminopyrimidine inhibitors of interleukin-1 receptor-associated kinase 4.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
7TD1
 
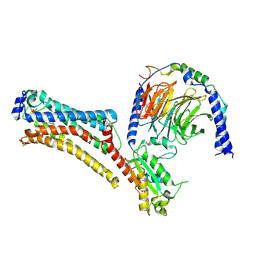 | | Lysophosphatidic acid receptor 1-Gi complex bound to LPA, state a | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, S, Paknejad, N, Zhu, L, Kihara, Y, Ray, D, Chun, J, Liu, W, Hite, R.K, Huang, X.Y. | | Deposit date: | 2021-12-30 | | Release date: | 2022-02-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Differential activation mechanisms of lipid GPCRs by lysophosphatidic acid and sphingosine 1-phosphate.
Nat Commun, 13, 2022
|
|
4XFY
 
 | |
7TED
 
 | | Human Ornithine Aminotransferase cocrystallized with its inhibitor, (S,E)-3-amino-4-(fluoromethylene)cyclopent-1-ene-1-carboxylate | | Descriptor: | (1S,3R,4S)-3-formyl-4-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]cyclopentane-1-carboxylic acid, Ornithine aminotransferase, mitochondrial, ... | | Authors: | Butrin, A, Zhu, W, Silverman, R, Liu, D. | | Deposit date: | 2022-01-04 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Rational Design, Synthesis, and Mechanism of (3 S ,4 R )-3-Amino-4-(difluoromethyl)cyclopent-1-ene-1-carboxylic Acid: Employing a Second-Deprotonation Strategy for Selectivity of Human Ornithine Aminotransferase over GABA Aminotransferase.
J.Am.Chem.Soc., 144, 2022
|
|
