6Y3E
 
 | |
4AF8
 
 | | The structural basis for metacaspase substrate specificity and activation | | Descriptor: | CHLORIDE ION, METACASPASE MCA2, SODIUM ION | | Authors: | McLuskey, K, Rudolf, J, Isaacs, N.W, Coombs, G.H, Moss, C.X, Mottram, J.C. | | Deposit date: | 2012-01-18 | | Release date: | 2012-05-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of a Trypanosoma Brucei Metacaspase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7SYV
 
 | | Structure of the wt IRES eIF5B-containing pre-48S initiation complex, open conformation. Structure 14(wt) | | Descriptor: | 18S rRNA, 40S ribosomal protein S2, 40S ribosomal protein S21, ... | | Authors: | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | Deposit date: | 2021-11-25 | | Release date: | 2022-07-20 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
7KU9
 
 | | The internal aldimine form of the wild-type Salmonella typhimurium Tryptophan Synthase with sodium ion at the metal coordination site, two molecules of F6F inhibitor at the enzyme alpha-site and another F6F molecule at the enzyme beta-site at 1.40 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-{[4-(TRIFLUOROMETHOXY)BENZOYL]AMINO}ETHYL DIHYDROGEN PHOSPHATE, CHLORIDE ION, ... | | Authors: | Hilario, E, Dunn, M.F, Mueller, L.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-12-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The internal aldimine form of the wild-type Salmonella typhimurium Tryptophan Synthase with sodium ion at the metal coordination site, two molecules of F6F inhibitor at the enzyme alpha-site and another F6F molecule at the enzyme beta-site at 1.40 Angstrom resolution.
To be Published
|
|
7OCS
 
 | | Mannitol-1-phosphate bound to the phosphatase domain of the bifunctional mannitol-1-phosphate dehydrogenase/phosphatase MtlD-D16A from Acinetobacter baumannii | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Tam, H.K, Mueller, V, Pos, K.M. | | Deposit date: | 2021-04-28 | | Release date: | 2022-04-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Unidirectional mannitol synthesis of Acinetobacter baumannii MtlD is facilitated by the helix-loop-helix-mediated dimer formation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4RQ6
 
 | |
6YXX
 
 | | State A of the Trypanosoma brucei mitoribosomal large subunit assembly intermediate | | Descriptor: | 12S ribosomal RNA, 50S ribosomal protein L13, putative, ... | | Authors: | Jaskolowski, M, Ramrath, D.J.F, Bieri, P, Niemann, M, Mattei, S, Calderaro, S, Leibundgut, M.A, Horn, E.K, Boehringer, D, Schneider, A, Ban, N. | | Deposit date: | 2020-05-04 | | Release date: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Insights into the Mechanism of Mitoribosomal Large Subunit Biogenesis.
Mol.Cell, 79, 2020
|
|
6Z5J
 
 | | Arrangement of the matrix protein M1 in influenza A/Hong Kong/1/1968 VLPs (HA,NA,M1,M2) | | Descriptor: | Matrix protein 1 | | Authors: | Peukes, J, Xiong, X, Erlendsson, S, Qu, K, Wan, W, Kraeusslich, H.-G, Briggs, J.A.G. | | Deposit date: | 2020-05-26 | | Release date: | 2020-10-14 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | The native structure of the assembled matrix protein 1 of influenza A virus.
Nature, 587, 2020
|
|
3WA3
 
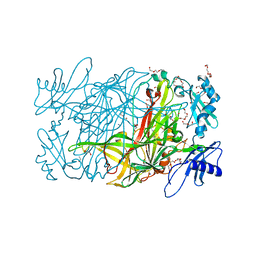 | | Crystal structure of copper amine oxidase from arthrobacter globiformis in N2 condition | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Murakawa, T, Hayashi, H, Sunami, T, Kurihara, K, Tamada, T, Kuroki, R, Suzuki, M, Tanizawa, K, Okajima, T. | | Deposit date: | 2013-04-22 | | Release date: | 2013-09-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | High-resolution crystal structure of copper amine oxidase from Arthrobacter globiformis: assignment of bound diatomic molecules as O2
Acta Crystallogr.,Sect.D, 69, 2013
|
|
5UJU
 
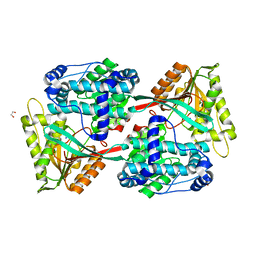 | |
3WXO
 
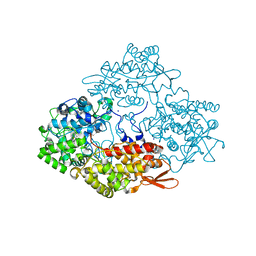 | | Crystal structure of isoniazid bound KatG catalase peroxidase from Synechococcus elongatus PCC7942 | | Descriptor: | Catalase-peroxidase, PROTOPORPHYRIN IX CONTAINING FE, SODIUM ION, ... | | Authors: | Wada, K, Tada, T, Kamachi, S. | | Deposit date: | 2014-08-04 | | Release date: | 2015-01-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | The crystal structure of isoniazid-bound KatG catalase-peroxidase from Synechococcus elongatus PCC7942.
Febs J., 282, 2015
|
|
6YUX
 
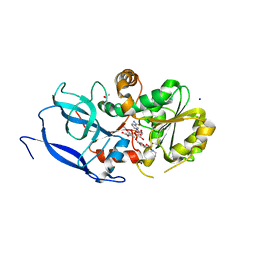 | | Crystal structure of Malus domestica Double Bond Reductase (MdDBR) ternary complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, ... | | Authors: | Caliandro, R, Polsinelli, I, Demitri, N, Benini, S. | | Deposit date: | 2020-04-27 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | The structural and functional characterization of Malus domestica double bond reductase MdDBR provides insights towards the identification of its substrates.
Int.J.Biol.Macromol., 171, 2021
|
|
6YXY
 
 | | State B of the Trypanosoma brucei mitoribosomal large subunit assembly intermediate | | Descriptor: | 12S ribosomal RNA, ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Jaskolowski, M, Ramrath, D.J.F, Bieri, P, Niemann, M, Mattei, S, Calderaro, S, Leibundgut, M.A, Horn, E.K, Boehringer, D, Schneider, A, Ban, N. | | Deposit date: | 2020-05-04 | | Release date: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural Insights into the Mechanism of Mitoribosomal Large Subunit Biogenesis.
Mol.Cell, 79, 2020
|
|
6ZIV
 
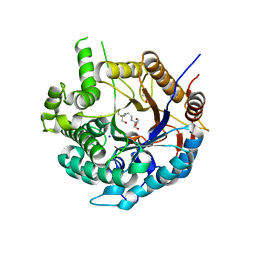 | | Crystal structure of a Beta-glucosidase from Alicyclobacillus acidiphilus | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-glucosidase, ... | | Authors: | Gourlay, L.J. | | Deposit date: | 2020-06-26 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Producing natural vanilla extract from green vanilla beans using a beta-glucosidase from Alicyclobacillus acidiphilus.
J.Biotechnol., 329, 2021
|
|
6Z5T
 
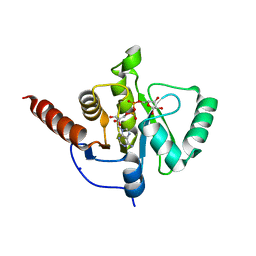 | | SARS-CoV-2 Macrodomain in complex with ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Replicase polyprotein 1ab, SODIUM ION | | Authors: | Zorzini, V, Rack, J, Ahel, I. | | Deposit date: | 2020-05-27 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.571 Å) | | Cite: | Viral macrodomains: a structural and evolutionary assessment of the pharmacological potential.
Open Biology, 10, 2020
|
|
8UGI
 
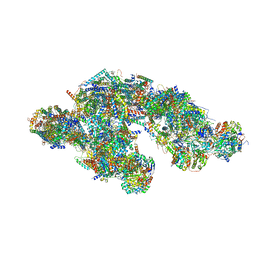 | | High resolution in-situ structure of typeA supercomplex in respiratory chain (I1III2IV1,composite) | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Zheng, W, Zhang, K, Zhu, J. | | Deposit date: | 2023-10-05 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | High-resolution in situ structures of mammalian respiratory supercomplexes.
Nature, 2024
|
|
6YT4
 
 | | Crystal structure of the N112A mutant of the light-driven sodium pump KR2 in the pentameric form, pH 8.0 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, EICOSANE, OLEIC ACID, ... | | Authors: | Kovalev, K, Maliar, N, Astashkin, R, Gordeliy, V. | | Deposit date: | 2020-04-23 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the N112A Mutant of the Light-Driven Sodium Pump KR2
Crystals, 2020
|
|
8UGR
 
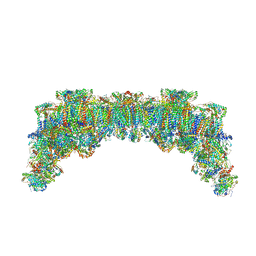 | | In-situ structure of typeX supercomplex in respiratory chain (composite) | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Zheng, W, Zhang, K, Zhu, J. | | Deposit date: | 2023-10-06 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | High-resolution in situ structures of mammalian respiratory supercomplexes.
Nature, 2024
|
|
8UGN
 
 | | In-situ structure of typeO supercomplex in respiratory chain (composite) | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Zheng, W, Zhang, K, Zhu, J. | | Deposit date: | 2023-10-05 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | High-resolution in situ structures of mammalian respiratory supercomplexes.
Nature, 2024
|
|
8UGJ
 
 | | In-situ structure of typeB supercomplex in respiratory chain (composite) | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Zheng, W, Zhang, K, Zhu, J. | | Deposit date: | 2023-10-05 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | High-resolution in situ structures of mammalian respiratory supercomplexes.
Nature, 2024
|
|
5SXX
 
 | |
5SYU
 
 | | Crystal structure of Burkholderia pseudomallei KatG E242Q variant | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Catalase-peroxidase, ... | | Authors: | Loewen, P.C. | | Deposit date: | 2016-08-12 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Catalase Activity of Catalase-Peroxidases Is Modulated by Changes in the pKa of the Distal Histidine.
Biochemistry, 56, 2017
|
|
5SXR
 
 | | Crystal structure of B. pseudomallei KatG with NAD bound | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Loewen, P.C. | | Deposit date: | 2016-08-10 | | Release date: | 2016-09-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Isonicotinic acid hydrazide conversion to Isonicotinyl-NAD by catalase-peroxidases.
J. Biol. Chem., 285, 2010
|
|
5T03
 
 | | Crystal structure of heparan sulfate 6-O-sulfotransferase with bound PAP and glucuronic acid containing hexasaccharide substrate | | Descriptor: | 1,2-ETHANEDIOL, 2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid, ADENOSINE-3'-5'-DIPHOSPHATE, ... | | Authors: | Pedersen, L.C, Moon, A.F, Krahn, J.M, Liu, J. | | Deposit date: | 2016-08-15 | | Release date: | 2017-02-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure Based Substrate Specificity Analysis of Heparan Sulfate 6-O-Sulfotransferases.
ACS Chem. Biol., 12, 2017
|
|
5SXQ
 
 | |
