6MGH
 
 | | X-ray structure of monomeric near-infrared fluorescent protein miRFP670nano | | Descriptor: | 3-[2-[(~{Z})-[5-[(~{Z})-[(3~{S},4~{R})-3-ethenyl-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-pyrrol-2-ylidene]methyl]-5-[(~{Z})-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Pletnev, S. | | Deposit date: | 2018-09-13 | | Release date: | 2018-12-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Smallest near-infrared fluorescent protein evolved from cyanobacteriochrome as versatile tag for spectral multiplexing.
Nat Commun, 10, 2019
|
|
6PJ4
 
 | |
7R23
 
 | |
6MIT
 
 | | LptBFGC from Enterobacter cloacae | | Descriptor: | CHLORIDE ION, LPS export ABC transporter permease LptF, Lipopolysaccharide export system ATP-binding protein, ... | | Authors: | Owens, T.W, Kahne, D, Kruse, A.C. | | Deposit date: | 2018-09-20 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of unidirectional export of lipopolysaccharide to the cell surface.
Nature, 567, 2019
|
|
7AJV
 
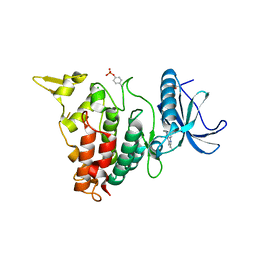 | | Structure of DYRK1A in complex with compound 38 | | Descriptor: | 4-(2,3-dibutylimidazo[4,5-b]pyridin-5-yl)pyridine-2,6-diamine, CHLORIDE ION, Dual specificity tyrosine-phosphorylation-regulated kinase 1A | | Authors: | Dokurno, P, Surgenor, A.E, Kotschy, A. | | Deposit date: | 2020-09-29 | | Release date: | 2021-05-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Guided Discovery of Potent and Selective DYRK1A Inhibitors.
J.Med.Chem., 64, 2021
|
|
6YQ6
 
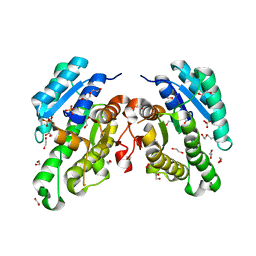 | | Promiscuous Reductase LugOII Catalyzes Keto-reduction at C1 during Lugdunomycin Biosynthesis | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Monooxygenase, ... | | Authors: | Xiao, X, Elsayed, S.S, Wu, C, van der Heul, H, Prota, A, Huang, J, Guo, R, Abrahams, J.P, van Wezel, G.P. | | Deposit date: | 2020-04-16 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Functional and Structural Insights into a Novel Promiscuous Ketoreductase of the Lugdunomycin Biosynthetic Pathway.
Acs Chem.Biol., 15, 2020
|
|
6Y68
 
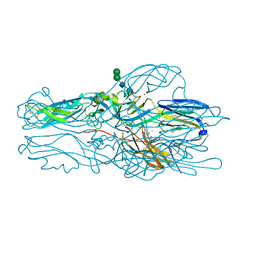 | |
8T35
 
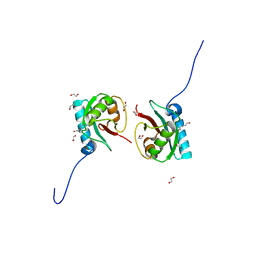 | | Crystal structure of K51 acetylated LC3A in complex with the LIR of TP53INP2/DOR | | Descriptor: | 1,2-ETHANEDIOL, Tumor protein p53-inducible nuclear protein 2,Microtubule-associated proteins 1A/1B light chain 3A | | Authors: | Ali, M.G.H, Wahba, H.M, Cyr, N, Omichinski, J.G. | | Deposit date: | 2023-06-07 | | Release date: | 2024-05-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Structural and functional characterization of the role of acetylation on the interactions of the human Atg8-family proteins with the autophagy receptor TP53INP2/DOR.
Autophagy, 20, 2024
|
|
7AQM
 
 | | ADP-ribosylserine hydrolase ARH3 of Latimeria chalumnae in complex with alpha-1''-O-methyl-ADP-ribose (meADPr) | | Descriptor: | ADP-ribosylhydrolase like 2, Adenosine 5'-diphosphoric acid beta-[(3beta,4beta-dihydroxy-5beta-methoxytetrahydrofuran-2alpha-yl)methyl] estere, MAGNESIUM ION | | Authors: | Rack, J.G.M, Zorzini, V, Ahel, I. | | Deposit date: | 2020-10-22 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanistic insights into the three steps of poly(ADP-ribosylation) reversal.
Nat Commun, 12, 2021
|
|
7AXN
 
 | | 14-3-3 sigma in complex with Pin1 binding site pS72 and covalently bound TCF521-026 | | Descriptor: | 14-3-3 protein sigma, 3-chloranyl-4-imidazol-1-yl-benzaldehyde, CALCIUM ION, ... | | Authors: | Wolter, M, Dijck, L.v, Ottmann, C. | | Deposit date: | 2020-11-10 | | Release date: | 2021-06-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
7AYF
 
 | | 14-3-3 sigma with Pin1 binding site pS72 and covalently bound TCF521-110 | | Descriptor: | 14-3-3 protein sigma, 6-(2-bromanylimidazol-1-yl)pyridine-3-carbaldehyde, CALCIUM ION, ... | | Authors: | Wolter, M, Dijck, L.v, Ottmann, C. | | Deposit date: | 2020-11-12 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
7AZ1
 
 | | 14-3-3 sigma with Pin1 binding site pS72 and covalently bound LvD1013 | | Descriptor: | 1-[4-methyl-3-(trifluoromethyl)phenyl]-2-phenyl-imidazole, 14-3-3 protein sigma, CALCIUM ION, ... | | Authors: | Wolter, M, Dijck, L.v, Ottmann, C. | | Deposit date: | 2020-11-14 | | Release date: | 2021-06-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
7R6V
 
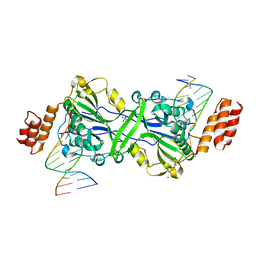 | | Human EXOG complexed with dRP-containing DNA | | Descriptor: | DNA (5'-D(*CP*GP*CP*AP*CP*GP*TP*CP*AP*GP*AP*A)-3'), DNA (5'-D(*CP*TP*GP*AP*CP*GP*TP*GP*CP*G)-3'), MAGNESIUM ION, ... | | Authors: | Szymanski, M.R, Yin, Y.W. | | Deposit date: | 2021-06-23 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Human EXOG possesses strong AP hydrolysis activity
To Be Published
|
|
7AZ2
 
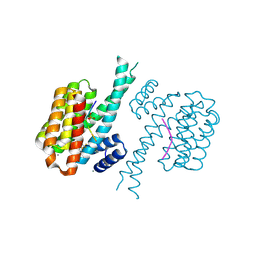 | | 14-3-3 sigma with Pin1 binding site pS72 and covalently bound LvD1014 | | Descriptor: | 1-[4-methyl-2-(trifluoromethyl)phenyl]-2-phenyl-imidazole, 14-3-3 protein sigma, CALCIUM ION, ... | | Authors: | Wolter, M, Dijck, L.v, Ottmann, C. | | Deposit date: | 2020-11-14 | | Release date: | 2021-06-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.081 Å) | | Cite: | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
7AOG
 
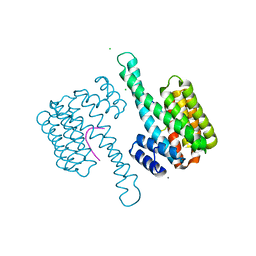 | | 14-3-3 sigma in complex with Pin1 binding site pS72 | | Descriptor: | 14-3-3 protein sigma, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Wolter, M, Dijck, L.v, Cossar, P.J, Ottmann, C. | | Deposit date: | 2020-10-14 | | Release date: | 2021-06-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
6POI
 
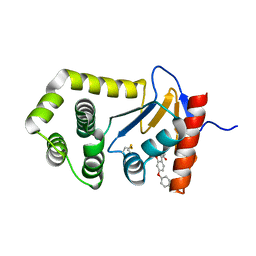 | | Crystal Structure of EcDsbA in complex phenyl ether 25 | | Descriptor: | (6-phenoxy-1-benzofuran-3-yl)acetic acid, COPPER (II) ION, Thiol:disulfide interchange protein DsbA | | Authors: | Ilyichova, O.V, Scanlon, M.J. | | Deposit date: | 2019-07-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | The Fragment-Based Development of a Benzofuran Hit as a New Class of Escherichia coli DsbA Inhibitors.
Molecules, 24, 2019
|
|
6POQ
 
 | | Crystal Structure of EcDsbA in complex with anisidine 16 | | Descriptor: | COPPER (II) ION, Thiol:disulfide interchange protein DsbA, {6-[(4-methoxyphenyl)amino]-1-benzofuran-3-yl}acetic acid | | Authors: | Ilyichova, O.V, Scanlon, M.J. | | Deposit date: | 2019-07-04 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Fragment-Based Development of a Benzofuran Hit as a New Class of Escherichia coli DsbA Inhibitors.
Molecules, 24, 2019
|
|
8T36
 
 | | Crystal structure of K49 acetylated LC3A in complex with the LIR of TP53INP2/DOR | | Descriptor: | 1,2-ETHANEDIOL, Tumor protein p53-inducible nuclear protein 2,Microtubule-associated proteins 1A/1B light chain 3A chimera | | Authors: | Ali, M.G.H, Wahba, H.M, Cyr, N, Omichinski, J.G. | | Deposit date: | 2023-06-07 | | Release date: | 2024-05-22 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.852 Å) | | Cite: | Structural and functional characterization of the role of acetylation on the interactions of the human Atg8-family proteins with the autophagy receptor TP53INP2/DOR.
Autophagy, 20, 2024
|
|
7AYR
 
 | |
8F7Z
 
 | | VRC34.01_mm28 bound to fusion peptide | | Descriptor: | HIV-1 Env Fusion Peptide, VRC34_m228 Light Chain, VRC34_mm28 Heavy Chain | | Authors: | Olia, A.S, Kwong, P.D. | | Deposit date: | 2022-11-21 | | Release date: | 2023-11-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Antibody-directed evolution reveals a mechanism for enhanced neutralization at the HIV-1 fusion peptide site.
Nat Commun, 14, 2023
|
|
7AX0
 
 | | Crystal structure of the computationally designed Scone-E protein co-crystallized with STA form a | | Descriptor: | Keggin (STA), PHOSPHATE ION, SconeE | | Authors: | Mylemans, B, Vandebroek, L, Parac-Vogt, T.N, Voet, A.R.D. | | Deposit date: | 2020-11-09 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of Scone: pseudosymmetric folding of a symmetric designer protein.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7AYQ
 
 | |
8T31
 
 | | Crystal structure of GABARAP in complex with the LIR of TP53INP2/DOR | | Descriptor: | Gamma-aminobutyric acid receptor-associated protein, Tumor protein p53-inducible nuclear protein 2 | | Authors: | Ali, M.G.H, Wahba, H.M, Cyr, N, Omichinski, J.G. | | Deposit date: | 2023-06-07 | | Release date: | 2024-05-22 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.096 Å) | | Cite: | Structural and functional characterization of the role of acetylation on the interactions of the human Atg8-family proteins with the autophagy receptor TP53INP2/DOR.
Autophagy, 20, 2024
|
|
7QZO
 
 | | Crystal structure of GacS D1 domain | | Descriptor: | CADMIUM ION, GLYCEROL, Histidine kinase | | Authors: | Fadel, F, Bassim, V, Botzanowski, T, Francis, V.I, Legrand, P, Porter, S.L, Bourne, Y, Cianferani, S, Vincent, F. | | Deposit date: | 2022-01-31 | | Release date: | 2022-07-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Insights into the atypical autokinase activity of the Pseudomonas aeruginosa GacS histidine kinase and its interaction with RetS.
Structure, 30, 2022
|
|
6MYZ
 
 | | Crystal structure of Tdp1 catalytic domain in complex with compound XZ520 | | Descriptor: | 1,2-ETHANEDIOL, 4-oxo-8-phenyl-1,4-dihydroquinoline-3-carboxylic acid, Tyrosyl-DNA phosphodiesterase 1 | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Burke Jr, T.R, Pommier, Y, Waugh, D.S. | | Deposit date: | 2018-11-02 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.661 Å) | | Cite: | Structure of Tdp1 catalytic domain in complex with compound XZ520
To Be Published
|
|
