8IF4
 
 | | Structure of human alpha-2/delta-1 without mirogabalin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Voltage-dependent calcium channel subunit alpha-2/delta-1 | | Authors: | Kozai, D, Numoto, N, Fujiyoshi, Y. | | Deposit date: | 2023-02-17 | | Release date: | 2023-04-05 | | Last modified: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Recognition Mechanism of a Novel Gabapentinoid Drug, Mirogabalin, for Recombinant Human alpha 2 delta 1, a Voltage-Gated Calcium Channel Subunit.
J.Mol.Biol., 435, 2023
|
|
8IF3
 
 | | Structure of human alpha-2/delta-1 with mirogabalin | | Descriptor: | 2-[(1R,5S,6S)-6-(aminomethyl)-3-ethyl-6-bicyclo[3.2.0]hept-3-enyl]acetic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kozai, D, Numoto, N, Fujiyoshi, Y. | | Deposit date: | 2023-02-17 | | Release date: | 2023-04-05 | | Last modified: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Recognition Mechanism of a Novel Gabapentinoid Drug, Mirogabalin, for Recombinant Human alpha 2 delta 1, a Voltage-Gated Calcium Channel Subunit.
J.Mol.Biol., 435, 2023
|
|
8IF2
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Omicron BQ.1.1 variant spike protein in complex with its receptor ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Kimura, K, Suzuki, T, Hashiguchi, T. | | Deposit date: | 2023-02-17 | | Release date: | 2023-05-17 | | Last modified: | 2023-05-24 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Convergent evolution of SARS-CoV-2 Omicron subvariants leading to the emergence of BQ.1.1 variant.
Nat Commun, 14, 2023
|
|
8IES
 
 | | Cryo-EM structure of ATP13A2 in the E1P-ADP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Polyamine-transporting ATPase 13A2, ... | | Authors: | Liu, Z.M, Mu, J.Q, Xue, C.Y. | | Deposit date: | 2023-02-15 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Conformational cycle of human polyamine transporter ATP13A2.
Nat Commun, 14, 2023
|
|
8IER
 
 | |
8IEO
 
 | | Cryo-EM structure of ATP13A2 in the nominal E1P state | | Descriptor: | MAGNESIUM ION, Polyamine-transporting ATPase 13A2, SPERMINE, ... | | Authors: | Liu, Z.M, Mu, J.Q, Xue, C.Y. | | Deposit date: | 2023-02-15 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Conformational cycle of human polyamine transporter ATP13A2.
Nat Commun, 14, 2023
|
|
8IEN
 
 | | Cryo-EM structure of ATP13A2 in the E2-Pi state | | Descriptor: | MAGNESIUM ION, Polyamine-transporting ATPase 13A2, SPERMINE, ... | | Authors: | Liu, Z.M, Mu, J.Q, Xue, C.Y. | | Deposit date: | 2023-02-15 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Conformational cycle of human polyamine transporter ATP13A2.
Nat Commun, 14, 2023
|
|
8IEM
 
 | | Cryo-EM structure of ATP13A2 in the E2P state | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, Polyamine-transporting ATPase 13A2, ... | | Authors: | Liu, Z.M, Mu, J.Q, Xue, C.Y. | | Deposit date: | 2023-02-15 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Conformational cycle of human polyamine transporter ATP13A2.
Nat Commun, 14, 2023
|
|
8IEL
 
 | |
8IEK
 
 | |
8IEJ
 
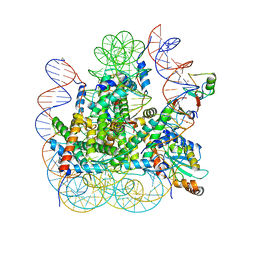 | | RNF20-RNF40/hRad6A-Ub/nucleosome complex | | Descriptor: | DNA (147-MER), E3 ubiquitin-protein ligase BRE1A, E3 ubiquitin-protein ligase BRE1B, ... | | Authors: | Ai, H, Deng, Z, Sun, M, Du, Y, Pan, M, Liu, L. | | Deposit date: | 2023-02-15 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Mechanistic insights into nucleosomal H2B monoubiquitylation mediated by yeast Bre1-Rad6 and its human homolog RNF20/RNF40-hRAD6A.
Mol.Cell, 83, 2023
|
|
8IEG
 
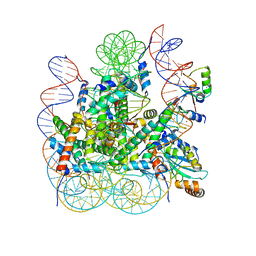 | | Bre1(mRBD-RING)/Rad6-Ub/nucleosome complex | | Descriptor: | DNA (147-MER), E3 ubiquitin-protein ligase BRE1, Histone H2A type 1-B/E, ... | | Authors: | Ai, H, Deng, Z, Pan, M, Liu, L. | | Deposit date: | 2023-02-15 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Mechanistic insights into nucleosomal H2B monoubiquitylation mediated by yeast Bre1-Rad6 and its human homolog RNF20/RNF40-hRAD6A.
Mol.Cell, 83, 2023
|
|
8IE2
 
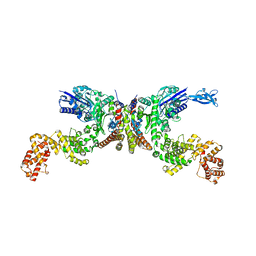 | | Crystal structure of Lactiplantibacillus plantarum GlyRS | | Descriptor: | Glycine--tRNA ligase alpha subunit, Glycine--tRNA ligase beta subunit | | Authors: | Yamashita, S, Tomita, K. | | Deposit date: | 2023-02-15 | | Release date: | 2023-06-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Mechanism of tRNA recognition by heterotetrameric glycyl-tRNA synthetase from lactic acid bacteria.
J.Biochem., 174, 2023
|
|
8IDU
 
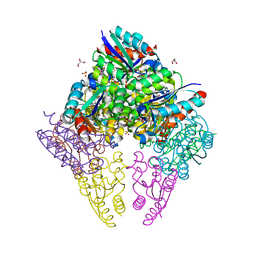 | | Crystal structure of substrate bound-form dehydroquinate dehydratase from Corynebacterium glutamicum | | Descriptor: | 1,3,4-TRIHYDROXY-5-OXO-CYCLOHEXANECARBOXYLIC ACID, 3-dehydroquinate dehydratase, GLYCEROL, ... | | Authors: | Lee, C.H, Kim, S, Kim, K.-J. | | Deposit date: | 2023-02-14 | | Release date: | 2023-12-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Biochemical Analysis of 3-Dehydroquinate Dehydratase from Corynebacterium glutamicum .
J Microbiol Biotechnol., 33, 2023
|
|
8IDQ
 
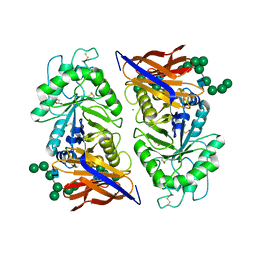 | | Crystal structure of reducing-end xylose-releasing exoxylanase in GH30 from Talaromyces cellulolyticus with xylose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Nakamichi, Y, Watanabe, M, Fujii, T, Inoue, H, Morita, T. | | Deposit date: | 2023-02-14 | | Release date: | 2023-05-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of reducing-end xylose-releasing exoxylanase in subfamily 7 of glycoside hydrolase family 30.
Proteins, 91, 2023
|
|
8IDP
 
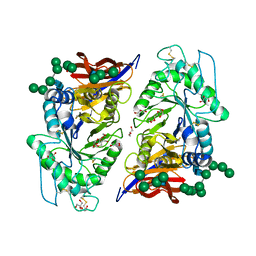 | | Crystal structure of reducing-end xylose-releasing exoxylanase in GH30 from Talaromyces cellulolyticus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Nakamichi, Y, Watanabe, M, Fujii, T, Inoue, H, Morita, T. | | Deposit date: | 2023-02-14 | | Release date: | 2023-05-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of reducing-end xylose-releasing exoxylanase in subfamily 7 of glycoside hydrolase family 30.
Proteins, 91, 2023
|
|
8IDN
 
 | | Cryo-EM structure of RBD/E77-Fab complex | | Descriptor: | E77 Fab heavy chain, E77 Fab light chain, Spike protein S1, ... | | Authors: | Lu, D.F, Zhang, Z.C. | | Deposit date: | 2023-02-13 | | Release date: | 2023-06-21 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | The structure of the RBD-E77 Fab complex reveals neutralization and immune escape of SARS-CoV-2.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8IDF
 
 | |
8IDE
 
 | | Structure of an ancient TsaD-TsaC-SUA5-TcdA modular enzyme (TsaN) | | Descriptor: | MANGANESE (II) ION, N(6)-L-threonylcarbamoyladenine synthase | | Authors: | Zhang, Z.L, Jin, M.Q, Yu, Z.J, Chen, W, Wang, X.L, Lei, D.S, Zhang, W.H. | | Deposit date: | 2023-02-13 | | Release date: | 2023-07-26 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structure-function analysis of an ancient TsaD-TsaC-SUA5-TcdA modular enzyme reveals a prototype of tRNA t6A and ct6A synthetases.
Nucleic Acids Res., 51, 2023
|
|
8IDA
 
 | | Overall structure of the LAT1-4F2hc bound with tyrosine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4F2 cell-surface antigen heavy chain, Large neutral amino acids transporter small subunit 1, ... | | Authors: | Yan, R.H, Li, Y.N, Shi, T.H. | | Deposit date: | 2023-02-12 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Overall structure of the LAT1-4F2hc bound with tyrosine
To Be Published
|
|
8ID8
 
 | | Cryo-EM structure of the TUG891 bound GPR120-Gi complex | | Descriptor: | 3-{4-[(4-fluoro-4'-methyl[1,1'-biphenyl]-2-yl)methoxy]phenyl}propanoic acid, Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Mao, C, Xiao, P, Tao, X, Qin, J, He, Q, Zhang, C, Yu, X, Zhang, Y, Sun, J. | | Deposit date: | 2023-02-12 | | Release date: | 2023-03-15 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Unsaturated bond recognition leads to biased signal in a fatty acid receptor.
Science, 380, 2023
|
|
8ID2
 
 | | Crystal structure of the ubiquitin-like domain in the SF3A1 subunit of human U2 snRNP complexed with the stem-loop 4 of U1 snRNA | | Descriptor: | RNA (5'-R(*GP*GP*GP*GP*AP*CP*UP*GP*CP*GP*UP*UP*CP*GP*CP*GP*CP*UP*UP*UP*CP*CP*CP*C)-3'), Splicing factor 3A subunit 1 | | Authors: | Terawaki, S, Nameki, N, Kuwasako, K. | | Deposit date: | 2023-02-12 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into recognition of SL4, the UUCG stem-loop, of human U1 snRNA by the ubiquitin-like domain, including the C-terminal tail in the SF3A1 subunit of U2 snRNP.
J.Biochem., 174, 2023
|
|
8ICZ
 
 | | DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF OF DATP (1 MILLIMOLAR), MNCL2 (5 MILLIMOLAR), AND LITHIUM SULFATE (75 MILLIMOLAR) | | Descriptor: | DNA (5'-D(*CP*AP*TP*TP*AP*GP*AP*A)-3'), DNA (5'-D(*TP*CP*TP*AP*AP*TP*G)-3'), PROTEIN (DNA POLYMERASE BETA (E.C.2.7.7.7)), ... | | Authors: | Pelletier, H, Sawaya, M.R. | | Deposit date: | 1996-01-04 | | Release date: | 1996-11-15 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Characterization of the metal ion binding helix-hairpin-helix motifs in human DNA polymerase beta by X-ray structural analysis.
Biochemistry, 35, 1996
|
|
8ICI
 
 | |
8ICH
 
 | |
