4N6O
 
 | | Crystal structure of reduced legumain in complex with cystatin E/M | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, IODIDE ION, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dall, E, Brandstetter, H. | | Deposit date: | 2013-10-14 | | Release date: | 2015-02-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and mechanism of an aspartimide-dependent Peptide ligase in human legumain.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
6E0M
 
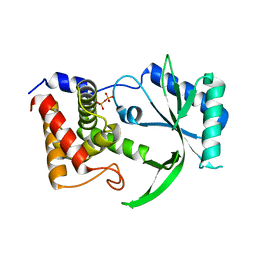 | | Structure of Elizabethkingia meningoseptica CdnE cyclic dinucleotide synthase | | Descriptor: | DIPHOSPHATE, cGAS/DncV-like nucleotidyltransferase in E. coli homolog | | Authors: | Eaglesham, J.B, Whiteley, A.T, de Oliveira Mann, C.C, Morehouse, B.R, Nieminen, E.A, King, D.S, Lee, A.S.Y, Mekalanos, J.J, Kranzusch, P.J. | | Deposit date: | 2018-07-06 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Bacterial cGAS-like enzymes synthesize diverse nucleotide signals.
Nature, 567, 2019
|
|
3F84
 
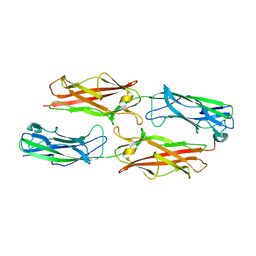 | |
1N03
 
 | | Model for Active RecA Filament | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, RecA protein | | Authors: | VanLoock, M.S, Yu, X, Yang, S, Lai, A.L, Low, C, Campbell, M.J, Egelman, E.H. | | Deposit date: | 2002-10-10 | | Release date: | 2003-02-25 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | ATP-Mediated Conformational Changes in the RecA Filament
Structure, 11, 2003
|
|
1N2Z
 
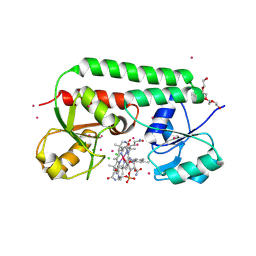 | | 2.0 Angstrom structure of BtuF, the vitamin B12 binding protein of E. coli | | Descriptor: | CADMIUM ION, CHLORIDE ION, CYANOCOBALAMIN, ... | | Authors: | Borths, E.L, Locher, K.P, Lee, A.T, Rees, D.C. | | Deposit date: | 2002-10-24 | | Release date: | 2002-12-18 | | Last modified: | 2021-08-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of Escherichia coli BtuF and binding to its cognate ATP binding cassette transporter
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
2G2R
 
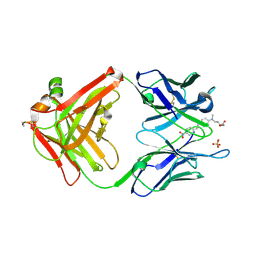 | |
4E4J
 
 | | Crystal structure of arginine deiminase from Mycoplasma penetrans | | Descriptor: | Arginine deiminase, CHLORIDE ION | | Authors: | Benach, J, Gallego, P, Planell, R, Querol, E, Perez Pons, J.A, Reverter, D. | | Deposit date: | 2012-03-13 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Characterization of the Enzymes Composing the Arginine Deiminase Pathway in Mycoplasma penetrans.
Plos One, 7, 2012
|
|
4QNL
 
 | | Crystal structure of tail fiber protein gp63.1 from E. coli phage G7C | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Riccio, C, Browning, C, Prokhorov, N, Letarov, A, Leiman, P.G. | | Deposit date: | 2014-06-18 | | Release date: | 2015-06-24 | | Last modified: | 2017-12-20 | | Method: | X-RAY DIFFRACTION (2.411 Å) | | Cite: | Function of bacteriophage G7C esterase tailspike in host cell adsorption.
Mol. Microbiol., 105, 2017
|
|
2OSE
 
 | | Crystal Structure of the Mimivirus Cyclophilin | | Descriptor: | CHLORIDE ION, Probable peptidyl-prolyl cis-trans isomerase | | Authors: | Eisenmesser, E.Z, Thai, V, Renesto, P, Raoult, D. | | Deposit date: | 2007-02-05 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural, biochemical, and in vivo characterization of the first virally encoded cyclophilin from the Mimivirus.
J.Mol.Biol., 378, 2008
|
|
4P1M
 
 | | The structure of Escherichia coli ZapA | | Descriptor: | CHLORIDE ION, Cell division protein ZapA | | Authors: | Kimber, M.S, Roach, E.J, Khursigara, C.M. | | Deposit date: | 2014-02-26 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure and Site-directed Mutational Analysis Reveals Key Residues Involved in Escherichia coli ZapA Function.
J.Biol.Chem., 289, 2014
|
|
3GLH
 
 | | Crystal Structure of the E. coli clamp loader bound to Psi Peptide | | Descriptor: | DNA polymerase III subunit delta, DNA polymerase III subunit delta', DNA polymerase III subunit tau | | Authors: | Kazmirski, S.L, Simonetta, K.R, Kuriyan, J. | | Deposit date: | 2009-03-12 | | Release date: | 2009-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.891 Å) | | Cite: | The mechanism of ATP-dependent primer-template recognition by a clamp loader complex.
Cell(Cambridge,Mass.), 137, 2009
|
|
3HG4
 
 | | Human alpha-galactosidase catalytic mechanism 3. Covalent intermediate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guce, A.I, Clark, N.E, Garman, S.C. | | Deposit date: | 2009-05-13 | | Release date: | 2009-11-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Catalytic mechanism of human alpha-galactosidase.
J.Biol.Chem., 285, 2010
|
|
3AX7
 
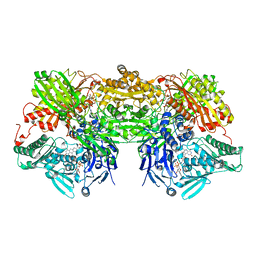 | | Bovine Xanthine Oxidase, protease cleaved form | | Descriptor: | 2-HYDROXYBENZOIC ACID, BICARBONATE ION, CALCIUM ION, ... | | Authors: | Ishikita, H, Eger, B.T, Pai, E.F, Okamoto, K, Nishino, T. | | Deposit date: | 2011-03-30 | | Release date: | 2012-02-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Protein conformational gating of enzymatic activity in xanthine oxidoreductase
J.Am.Chem.Soc., 134, 2012
|
|
3HG2
 
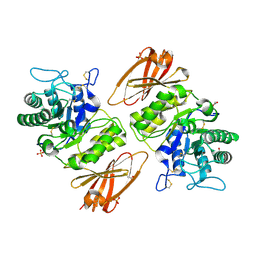 | | Human alpha-galactosidase catalytic mechanism 1. Empty active site | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID, Alpha-galactosidase A, ... | | Authors: | Guce, A.I, Clark, N.E, Garman, S.C. | | Deposit date: | 2009-05-13 | | Release date: | 2009-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Catalytic mechanism of human alpha-galactosidase.
J.Biol.Chem., 285, 2010
|
|
3AX9
 
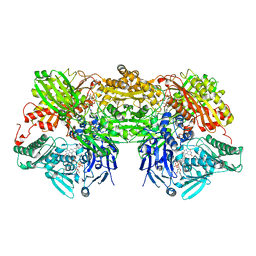 | | Bovine xanthine oxidase, protease cleaved form | | Descriptor: | 2-HYDROXYBENZOIC ACID, BICARBONATE ION, CALCIUM ION, ... | | Authors: | Ishikita, H, Eger, B.T, Pai, E.F, Okamoto, K, Nishino, T. | | Deposit date: | 2011-03-31 | | Release date: | 2012-02-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Protein conformational gating of enzymatic activity in xanthine oxidoreductase
J.Am.Chem.Soc., 134, 2012
|
|
1TPH
 
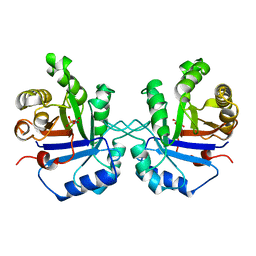 | | 1.8 ANGSTROMS CRYSTAL STRUCTURE OF WILD TYPE CHICKEN TRIOSEPHOSPHATE ISOMERASE-PHOSPHOGLYCOLOHYDROXAMATE COMPLEX | | Descriptor: | PHOSPHOGLYCOLOHYDROXAMIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Zhang, Z, Sugio, S, Komives, E.A, Liu, K.D, Knowles, J.R, Petsko, G.A, Ringe, D. | | Deposit date: | 1993-12-22 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of recombinant chicken triosephosphate isomerase-phosphoglycolohydroxamate complex at 1.8-A resolution.
Biochemistry, 33, 1994
|
|
3HG3
 
 | | Human alpha-galactosidase catalytic mechanism 2. Substrate bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-galactosidase A, ... | | Authors: | Guce, A.I, Clark, N.E, Garman, S.C. | | Deposit date: | 2009-05-13 | | Release date: | 2009-11-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Catalytic mechanism of human alpha-galactosidase.
J.Biol.Chem., 285, 2010
|
|
5FEQ
 
 | | EGFR KINASE DOMAIN IN COMPLEX WITH A COVALENT AMINOBENZIMIDAZOLE | | Descriptor: | Epidermal growth factor receptor, ~{N}-[1-[(3~{R})-1-[4-(dimethylamino)but-2-enoyl]azepan-3-yl]-7-methyl-benzimidazol-2-yl]-2-methyl-pyridine-4-carboxamide | | Authors: | DiDonato, M, Spraggon, G. | | Deposit date: | 2015-12-17 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Discovery of (R,E)-N-(7-Chloro-1-(1-[4-(dimethylamino)but-2-enoyl]azepan-3-yl)-1H-benzo[d]imidazol-2-yl)-2-methylisonicotinamide (EGF816), a Novel, Potent, and WT Sparing Covalent Inhibitor of Oncogenic (L858R, ex19del) and Resistant (T790M) EGFR Mutants for the Treatment of EGFR Mutant Non-Small-Cell Lung Cancers.
J.Med.Chem., 59, 2016
|
|
1Y52
 
 | | structure of insect cell (Baculovirus) expressed AVR4 (C122S)-biotin complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Avidin-related protein 4/5, BIOTIN | | Authors: | Eisenberg-Domovich, Y, Hytonen, V.P, Wilchek, M, Bayer, E.A, Kulomaa, M.S, Livnah, O. | | Deposit date: | 2004-12-02 | | Release date: | 2005-05-24 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High-resolution crystal structure of an avidin-related protein: insight into high-affinity biotin binding and protein stability.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
3IGN
 
 | | Crystal Structure of the GGDEF domain from Marinobacter aquaeolei diguanylate cyclase complexed with c-di-GMP - Northeast Structural Genomics Consortium Target MqR89a | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Diguanylate cyclase | | Authors: | Vorobiev, S, Neely, H, Seetharaman, J, Wang, H, Foote, E.L, Ciccosanti, C, Sahdev, S, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-07-28 | | Release date: | 2009-08-11 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structure of a catalytically active GG(D/E)EF diguanylate cyclase domain from Marinobacter aquaeolei with bound c-di-GMP product.
J.Struct.Funct.Genom., 13, 2012
|
|
3B4N
 
 | | Crystal Structure Analysis of Pectate Lyase PelI from Erwinia chrysanthemi | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Endo-pectate lyase, ... | | Authors: | Creze, C, Castang, S, Derivery, E, Haser, R, Shevchik, V, Gouet, P. | | Deposit date: | 2007-10-24 | | Release date: | 2008-04-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Crystal Structure of Pectate Lyase PelI from Soft Rot Pathogen Erwinia chrysanthemi in Complex with Its Substrate.
J.Biol.Chem., 283, 2008
|
|
1UHV
 
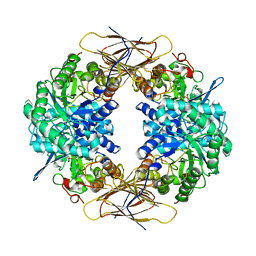 | | Crystal structure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase | | Descriptor: | 1,5-anhydro-2-deoxy-2-fluoro-D-xylitol, Beta-xylosidase | | Authors: | Yang, J.K, Yoon, H.J, Ahn, H.J, Il Lee, B, Pedelacq, J.D, Liong, E.C, Berendzen, J, Laivenieks, M, Vieille, C, Zeikus, G.J, Vocadlo, D.J, Withers, S.G, Suh, S.W. | | Deposit date: | 2003-07-11 | | Release date: | 2003-12-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase.
J.Mol.Biol., 335, 2004
|
|
2EWF
 
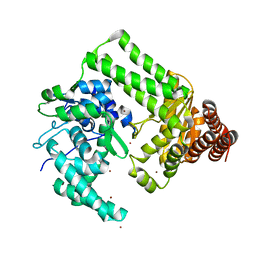 | | Crystal structure of the site-specific DNA nickase N.BspD6I | | Descriptor: | BROMIDE ION, Nicking endonuclease N.BspD6I | | Authors: | Kachalova, G.S, Bartunik, H.D, Artyukh, R.I, Rogulin, E.A, Perevyazova, T.A, Zheleznaya, L.A, Matvienko, N.I. | | Deposit date: | 2005-11-03 | | Release date: | 2006-11-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural analysis of the heterodimeric type IIS restriction endonuclease R.BspD6I acting as a complex between a monomeric site-specific nickase and a catalytic subunit.
J.Mol.Biol., 384, 2008
|
|
3HJ0
 
 | |
1YZ6
 
 | |
