9C8O
 
 | |
5NM4
 
 | | A2A Adenosine receptor room-temperature structure determined by serial femtosecond crystallography | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, CHOLESTEROL, ... | | Authors: | Weinert, T, Cheng, R, James, D, Gashi, D, Nogly, P, Jaeger, K, Hennig, M, Standfuss, J. | | Deposit date: | 2017-04-05 | | Release date: | 2017-09-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Serial millisecond crystallography for routine room-temperature structure determination at synchrotrons.
Nat Commun, 8, 2017
|
|
8V9F
 
 | | BRD4 BD1 liganded with macrocyclic compound 2d (JJ-II-363A) | | Descriptor: | (4bS)-1-ethyl-7,20-dimethyl-4b,10,11,21-tetrahydro-2H,17H-dibenzo[12',13':5',6'][1,4,8]trioxacyclotridecino[11',10':4,5]pyrido[2,3-d]pyrimidine-2,4,19(1H,3H)-trione, 1,2-ETHANEDIOL, Bromodomain-containing protein 4 | | Authors: | Schonbrunn, E, Chan, A. | | Deposit date: | 2023-12-08 | | Release date: | 2024-12-18 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Macrocyclic dihydropyridine analogs as pan-BET BD2-preferred inhibitors.
Eur.J.Med.Chem., 290, 2025
|
|
5LBZ
 
 | | Structure of the human quinone reductase 2 (NQO2) in complex with pacritinib | | Descriptor: | 11-(2-pyrrolidin-1-yl-ethoxy)-14,19-dioxa-5,7,26-triaza-tetracyclo[19.3.1.1(2,6).1(8,12)]heptacosa-1(25),2(26),3,5,8,10,12(27),16,21,23-decaene, FLAVIN-ADENINE DINUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Schneider, S, Medard, G, Kuster, B. | | Deposit date: | 2016-06-17 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The target landscape of clinical kinase drugs.
Science, 358, 2017
|
|
6ZAW
 
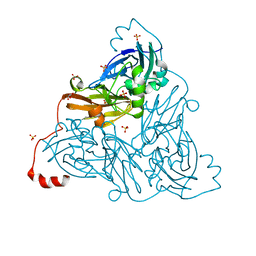 | | Damage-free NO-bound copper nitrite reductase from Bradyrhizobium sp. ORS 375 (two-domain) determined by serial femtosecond rotation crystallography | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Rose, S.L, Antonyuk, S.V, Sasaki, D, Yamashita, K, Hirata, K, Ueno, G, Ago, H, Eady, R.R, Tosha, T, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2020-06-05 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | An unprecedented insight into the catalytic mechanism of copper nitrite reductase from atomic-resolution and damage-free structures.
Sci Adv, 7, 2021
|
|
6H9Z
 
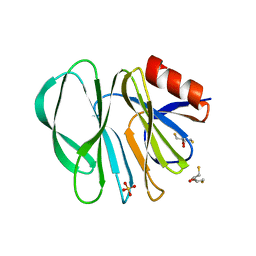 | | Molecular bases of histo-blood group antigen recognition by the most common human rotavirus | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Outer capsid protein VP4, SULFATE ION | | Authors: | Ciges-Tomas, J.R, Gozalbo-Rovira, R, Vila-Vicent, S, Buesa, J, Santiso-Bellon, C, Monedero, V, Yebra, M.J, Rodriguez-Diaz, J, Marina, A. | | Deposit date: | 2018-08-06 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Unraveling the role of the secretor antigen in human rotavirus attachment to histo-blood group antigens.
Plos Pathog., 15, 2019
|
|
6FIU
 
 | |
9NMO
 
 | |
9NMR
 
 | |
6ZAX
 
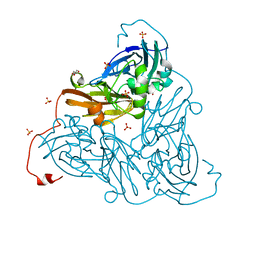 | | Nitrite-bound copper nitrite reductase from Bradyrhizobium sp. ORS 375 (two-domain) at low dose (0.5 MGy) | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, GLYCEROL, ... | | Authors: | Rose, S.L, Antonyuk, S.V, Sasaki, D, Yamashita, K, Hirata, K, Ueno, G, Ago, H, Eady, R.R, Tosha, T, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2020-06-05 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | An unprecedented insight into the catalytic mechanism of copper nitrite reductase from atomic-resolution and damage-free structures.
Sci Adv, 7, 2021
|
|
7YOR
 
 | | Recj2 and dCMP complex from Methanocaldococcus jannaschii | | Descriptor: | 1,2-ETHANEDIOL, [5-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-3-oxidanyl-furan-2-yl]methyl dihydrogen phosphate, archaeal nuclease RecJ2 | | Authors: | Wang, W.W, Liu, X.P. | | Deposit date: | 2022-08-02 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | The structure of nuclease RecJ2 from Methanocaldococcus jannaschii
To Be Published
|
|
2W90
 
 | | Geobacillus stearothermophilus 6-phosphogluconate dehydrogenase with bound 6- phosphogluconate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-PHOSPHOGLUCONATE DEHYDROGENASE, DECARBOXYLATING, ... | | Authors: | Cameron, S, Martini, V.P, Iulek, J, Hunter, W.N. | | Deposit date: | 2009-01-20 | | Release date: | 2009-04-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Geobacillus Stearothermophilus 6-Phosphogluconate Dehydrogenase, Complexed with 6-Phosphogluconate.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
5YJL
 
 | | Crystal structure of Arabidopsis glutamyl-tRNA reductase in complex with NADPH and GBP | | Descriptor: | Glutamyl-tRNA reductase 1, Glutamyl-tRNA reductase-binding protein, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhao, A, Han, F. | | Deposit date: | 2017-10-11 | | Release date: | 2018-05-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of Arabidopsis thaliana glutamyl-tRNAGlureductase in complex with NADPH and glutamyl-tRNAGlureductase binding protein
Photosyn. Res., 137, 2018
|
|
8BJL
 
 | |
6EPT
 
 | | The ATAD2 bromodomain in complex with compound 12 | | Descriptor: | (2~{R})-~{N}-[5-(5-azanylpyridin-3-yl)-4-ethanoyl-1,3-thiazol-2-yl]piperazine-2-carboxamide, (2~{S})-~{N}-[5-(5-azanylpyridin-3-yl)-4-ethanoyl-1,3-thiazol-2-yl]piperazine-2-carboxamide, ATPase family AAA domain-containing protein 2, ... | | Authors: | Sledz, P, Caflisch, A. | | Deposit date: | 2017-10-12 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Hitting a Moving Target: Simulation and Crystallography Study of ATAD2 Bromodomain Blockers.
Acs Med.Chem.Lett., 11, 2020
|
|
5JH6
 
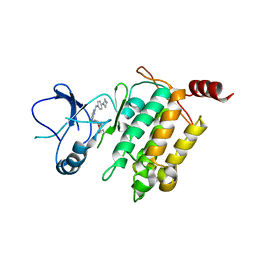 | | Crystal structure of TL10-92 bound to TAK1-TAB1 | | Descriptor: | 2-[(5-chloro-2-{[4-(4-methylpiperazin-1-yl)phenyl]amino}pyrimidin-4-yl)amino]phenyl prop-2-enoate, Mitogen-activated protein kinase kinase kinase 7,TGF-beta-activated kinase 1 and MAP3K7-binding protein 1 | | Authors: | Gurbani, D, Westover, K.D. | | Deposit date: | 2016-04-20 | | Release date: | 2017-02-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.365 Å) | | Cite: | Structure-guided development of covalent TAK1 inhibitors.
Bioorg. Med. Chem., 25, 2017
|
|
8YJN
 
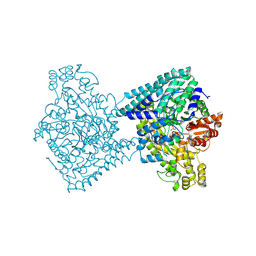 | | Structure of E. coli glycyl radical enzyme YbiW with bound glycerol | | Descriptor: | GLYCEROL, Probable dehydratase YbiW | | Authors: | Xue, B, Wei, Y, Robinson, R.C, Yew, W.S, Zhang, Y. | | Deposit date: | 2024-03-02 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | A Widespread Radical-Mediated Glycolysis Pathway.
J.Am.Chem.Soc., 146, 2024
|
|
8PYA
 
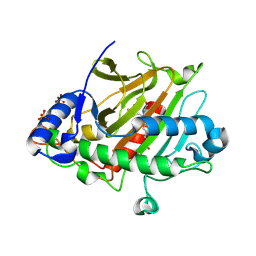 | | Isopenicillin N synthase in complex with Fe, O2, ACdV or thioaldehyde after 4s O2 exposure | | Descriptor: | (2S)-2-azanyl-6-[[(2R)-1-[[(2R)-3-methyl-1-oxidanyl-1-oxidanylidene-butan-2-yl]amino]-1-oxidanylidene-3-sulfanylidene-propan-2-yl]amino]-6-oxidanylidene-hexanoic acid, FE (III) ION, Isopenicillin N synthase, ... | | Authors: | Rabe, P, Schofield, C.J. | | Deposit date: | 2023-07-25 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Isopenicillin N synthase in complex with Fe, O2, ACdV or thioaldehyde after 4s O2 exposure
To Be Published
|
|
6FNU
 
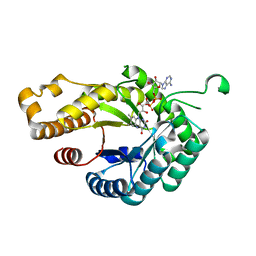 | | Structure of S. cerevisiae Methylenetetrahydrofolate reductase 1, catalytic domain | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Methylenetetrahydrofolate reductase 1 | | Authors: | Kopec, J, Rembeza, E, Bezerra, G.A, Newman, J, Bountra, C, Froese, D.S, Baumgartner, M, Yue, W.W, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-05 | | Release date: | 2018-03-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural basis for the regulation of human 5,10-methylenetetrahydrofolate reductase by phosphorylation and S-adenosylmethionine inhibition.
Nat Commun, 9, 2018
|
|
5KVS
 
 | | Substrate Analog and NADP+ bound structure of Irp3, a Thiazolinyl Imine Reductase from Yersinia enterocolitica | | Descriptor: | (4~{R})-2-[2-(2-hydroxyphenyl)-1,3-thiazol-4-yl]-4,5-dihydro-1,3-thiazole-4-carboxylic acid, 1,2-ETHANEDIOL, FORMIC ACID, ... | | Authors: | Meneely, K.M, Lamb, A.L. | | Deposit date: | 2016-07-15 | | Release date: | 2016-09-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Holo Structure and Steady State Kinetics of the Thiazolinyl Imine Reductases for Siderophore Biosynthesis.
Biochemistry, 55, 2016
|
|
4Z15
 
 | | MIF in complex with 3-(2-furylmethyl)-2-thioxo-1,3-thiazolan-4-one | | Descriptor: | ISOPROPYL ALCOHOL, Macrophage migration inhibitory factor, SULFATE ION | | Authors: | Cho, T.Y. | | Deposit date: | 2015-03-26 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for decreased induction of class IB PI3-kinases expression by MIF inhibitors.
J. Cell. Mol. Med., 21, 2017
|
|
8VK3
 
 | |
9QDI
 
 | | Crystal structure of BF3526 peptidase from Bacteroides fragilis in complex with a peptide | | Descriptor: | 1,2-ETHANEDIOL, 1,4-BUTANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Martinez Gascuena, A, Marquez-Monino, M.A, Manzanares-Gomez, A, Aguillo-Urarte, M, Trastoy, B. | | Deposit date: | 2025-03-06 | | Release date: | 2025-07-16 | | Last modified: | 2025-09-10 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Molecular basis of Fab-dependent IgA antibody recognition by gut-bacterial metallopeptidases.
Embo J., 44, 2025
|
|
8VJK
 
 | | Structure of mouse RyR1 (high-Ca2+/CFF/ATP dataset) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, ... | | Authors: | Weninger, G, Marks, A.R. | | Deposit date: | 2024-01-07 | | Release date: | 2024-01-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Structural insights into the regulation of RyR1 by S100A1.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8VJJ
 
 | | Structure of mouse RyR1 (EGTA-only dataset) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Peptidyl-prolyl cis-trans isomerase FKBP1A, Ryanodine receptor 1, ... | | Authors: | Weninger, G, Marks, A.R. | | Deposit date: | 2024-01-07 | | Release date: | 2024-01-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Structural insights into the regulation of RyR1 by S100A1.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
