4ZBT
 
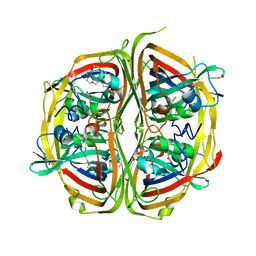 | | Streptomyces bingchenggensis aldolase-dehydratase in Schiff base complex with pyruvate | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Acetoacetate decarboxylase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Mydy, L.S, Silvaggi, N.R. | | Deposit date: | 2015-04-15 | | Release date: | 2015-06-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Sbi00515, a Protein of Unknown Function from Streptomyces bingchenggensis, Highlights the Functional Versatility of the Acetoacetate Decarboxylase Scaffold.
Biochemistry, 54, 2015
|
|
3OJX
 
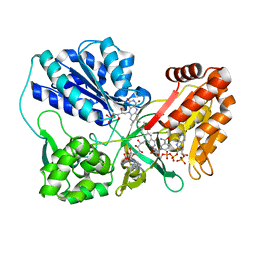 | | Disulfide crosslinked cytochrome P450 reductase inactive | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Xia, C, Hamdane, D, Shen, A, Choi, V, Kasper, C, Zhang, H, Im, S.-C, Waskell, L, Kim, J.-J.P. | | Deposit date: | 2010-08-23 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Conformational Changes of NADPH-Cytochrome P450 Oxidoreductase Are Essential for Catalysis and Cofactor Binding.
J.Biol.Chem., 286, 2011
|
|
3OR0
 
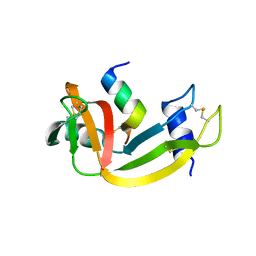 | |
3PX2
 
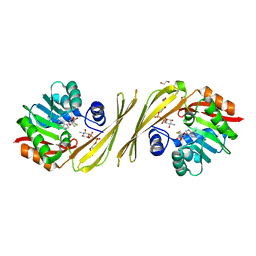 | | Structure of TylM1 from Streptomyces fradiae H123N mutant in complex with SAH and dTDP-Quip3N | | Descriptor: | 1,2-ETHANEDIOL, N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Holden, H.M, Carney, A.E. | | Deposit date: | 2010-12-09 | | Release date: | 2010-12-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Molecular Architecture of TylM1 from Streptomyces fradiae: An N,N-Dimethyltransferase Involved in the Production of dTDP-d-mycaminose .
Biochemistry, 50, 2011
|
|
3Q15
 
 | | Crystal Structure of RapH complexed with Spo0F | | Descriptor: | GLYCEROL, MAGNESIUM ION, Response regulator aspartate phosphatase H, ... | | Authors: | Parashar, V, Neiditch, M.B. | | Deposit date: | 2010-12-16 | | Release date: | 2011-02-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.192 Å) | | Cite: | Structural basis of response regulator dephosphorylation by Rap phosphatases.
Plos Biol., 9, 2011
|
|
3PI6
 
 | |
3OJW
 
 | | Disulfide crosslinked cytochrome P450 reductase inactive | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADPH-cytochrome P450 reductase | | Authors: | Xia, C, Hamdane, D, Shen, A, Choi, V, Kasper, C, Zhang, H, Im, S.-C, Waskell, L, Kim, J.-J.P. | | Deposit date: | 2010-08-23 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformational Changes of NADPH-Cytochrome P450 Oxidoreductase Are Essential for Catalysis and Cofactor Binding.
J.Biol.Chem., 286, 2011
|
|
1D0T
 
 | |
1D0U
 
 | |
3Q1X
 
 | |
3N30
 
 | | Crystal Structure of cubic Zn3-hUb (human ubiquitin) adduct | | Descriptor: | Ubiquitin, ZINC ION | | Authors: | Siliqi, D, Caliandro, R, Arnesano, F, Natile, G, Falini, G, Fermani, S, Belviso, B.D. | | Deposit date: | 2010-05-19 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystallographic analysis of metal-ion binding to human ubiquitin.
Chemistry, 17, 2011
|
|
3N32
 
 | | The crystal structure of human Ubiquitin adduct with Zeise's salt | | Descriptor: | PLATINUM (II) ION, Ubiquitin | | Authors: | Siliqi, D, Caliandro, R, Falini, G, Fermani, S, Natile, G, Arnesano, F, Belviso, B.D. | | Deposit date: | 2010-05-19 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.795 Å) | | Cite: | Crystallographic analysis of metal-ion binding to human ubiquitin.
Chemistry, 17, 2011
|
|
3MXG
 
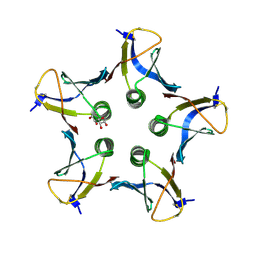 | | Structure of Shiga Toxin type 2 (Stx2) B Pentamer Mutant Q40L | | Descriptor: | D-xylose, Shiga-like toxin 2 subunit B | | Authors: | Kovall, R.A, Vander Wielen, B.D, Friedmann, D.R, Flagler, M.J. | | Deposit date: | 2010-05-07 | | Release date: | 2011-01-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Molecular basis of differential B-pentamer stability of Shiga toxins 1 and 2.
Plos One, 5, 2010
|
|
3N4Z
 
 | |
4Z84
 
 | | PKAB3 in complex with pyrrolidine inhibitor 34a | | Descriptor: | 7-[(3S,4R)-4-(3-chlorophenyl)carbonylpyrrolidin-3-yl]-3H-quinazolin-4-one, METHANOL, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Lund, B.A, Alam, K.A, Engh, R.A. | | Deposit date: | 2015-04-08 | | Release date: | 2015-12-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.554 Å) | | Cite: | Addressing the Glycine-Rich Loop of Protein Kinases by a Multi-Facetted Interaction Network: Inhibition of PKA and a PKB Mimic.
Chemistry, 22, 2016
|
|
4ZBO
 
 | | Streptomyces bingchenggensis acetoacetate decarboxylase in non-covalent complex with potassium formate | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Acetoacetate decarboxylase, ... | | Authors: | Mydy, L.S, Silvaggi, N.R. | | Deposit date: | 2015-04-15 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Sbi00515, a Protein of Unknown Function from Streptomyces bingchenggensis, Highlights the Functional Versatility of the Acetoacetate Decarboxylase Scaffold.
Biochemistry, 54, 2015
|
|
4ZGM
 
 | | Crystal structure of Semaglutide peptide backbone in complex with the GLP-1 receptor extracellular domain | | Descriptor: | 3,6,9,12,15,18-hexaoxahexacosan-1-ol, Glucagon-like peptide 1 receptor, Semaglutide peptide backbone; 8Aib,34R-GLP-1(7-37)-OH | | Authors: | Reedtz-Runge, S. | | Deposit date: | 2015-04-23 | | Release date: | 2015-09-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of the Once-Weekly Glucagon-Like Peptide-1 (GLP-1) Analogue Semaglutide.
J.Med.Chem., 58, 2015
|
|
3Q3B
 
 | |
3NEQ
 
 | | Crystal structure of the chimeric muscarinic toxin MT7 with loop 3 from MT1 | | Descriptor: | SULFATE ION, Three-finger muscarinic toxin 7 | | Authors: | Stura, E.A, Servent, D, Menez, R, Mournier, G, Menez, A, Fruchart-Gaillard, C. | | Deposit date: | 2010-06-09 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Engineering of three-finger fold toxins creates ligands with original pharmacological profiles for muscarinic and adrenergic receptors.
Plos One, 7, 2012
|
|
4Z83
 
 | | PKAB3 in complex with pyrrolidine inhibitor 47a | | Descriptor: | 7-{(3S,4R)-4-[(5-bromothiophen-2-yl)carbonyl]pyrrolidin-3-yl}quinazolin-4(3H)-one, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Lund, B.A, Alam, K.A, Engh, R.A. | | Deposit date: | 2015-04-08 | | Release date: | 2015-12-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Addressing the Glycine-Rich Loop of Protein Kinases by a Multi-Facetted Interaction Network: Inhibition of PKA and a PKB Mimic.
Chemistry, 22, 2016
|
|
3NAN
 
 | | SR Ca(2+)-ATPase in the HnE2 state complexed with a Thapsigargin derivative Boc-(phi)Tg | | Descriptor: | (3S,3aR,4S,6S,6aR,7S,8S,9R,9aS,9bS)-6-(acetyloxy)-4-{[4-(3-{6-[(tert-butoxycarbonyl)amino]hexyl}-4-hydroxyphenyl)butanoyl]oxy}-3,3a-dihydroxy-3,6,9-trimethyl-8-{[(2Z)-2-methylbut-2-enoyl]oxy}-2-oxododecahydroazuleno[4,5-b]furan-7-yl octanoate, MAGNESIUM ION, PHOSPHATIDYLETHANOLAMINE, ... | | Authors: | Winther, A.M.L, Sonntag, Y, Olesen, C, Moller, J.V, Nissen, P. | | Deposit date: | 2010-06-02 | | Release date: | 2010-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Critical roles of hydrophobicity and orientation of side chains for inactivation of sarcoplasmic reticulum Ca2+-ATPase with thapsigargin and thapsigargin analogs
J.Biol.Chem., 285, 2010
|
|
3PX3
 
 | | Structure of TylM1 from Streptomyces fradiae H123A mutant in complex with SAH and dTDP-Quip3N | | Descriptor: | 1,2-ETHANEDIOL, N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Holden, H.M, Carney, A.E. | | Deposit date: | 2010-12-09 | | Release date: | 2010-12-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Architecture of TylM1 from Streptomyces fradiae: An N,N-Dimethyltransferase Involved in the Production of dTDP-d-mycaminose .
Biochemistry, 50, 2011
|
|
3NHN
 
 | | Crystal structure of the SRC-family kinase HCK SH3-SH2-linker regulatory region | | Descriptor: | Tyrosine-protein kinase HCK | | Authors: | Alvarado, J.J, Betts, L, Moroco, J.A, Smithgall, T.E, Yeh, J.I. | | Deposit date: | 2010-06-14 | | Release date: | 2010-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal Structure of the Src Family Kinase Hck SH3-SH2 Linker Regulatory Region Supports an SH3-dominant Activation Mechanism.
J.Biol.Chem., 285, 2010
|
|
3NAM
 
 | | SR Ca(2+)-ATPase in the HnE2 state complexed with the Thapsigargin derivative dOTg | | Descriptor: | (3S,3aR,4S,6S,6aS,8R,9R,9aR,9bS)-6-(acetyloxy)-4-(butanoyloxy)-3,3a-dihydroxy-3,6,9-trimethyl-2-oxododecahydroazuleno[4,5-b]furan-8-yl (2Z)-2-methylbut-2-enoate, MAGNESIUM ION, PHOSPHATIDYLETHANOLAMINE, ... | | Authors: | Winther, A.M.L, Sonntag, Y, Olesen, C, Moller, J.V, Nissen, P. | | Deposit date: | 2010-06-02 | | Release date: | 2010-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Critical roles of hydrophobicity and orientation of side chains for inactivation of sarcoplasmic reticulum Ca2+-ATPase with thapsigargin and thapsigargin analogs
J.Biol.Chem., 285, 2010
|
|
3N4Y
 
 | |
