3B69
 
 | | T cruzi Trans-sialidase complex with benzoylated NANA derivative | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-acetamido-9-(benzoylamino)-3,5,9-trideoxy-3-fluoro-D-erythro-alpha-L-manno-non-2-ulopyranosonic acid, CHLORIDE ION, ... | | Authors: | Buschiazzo, A. | | Deposit date: | 2007-10-28 | | Release date: | 2008-05-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | A new generation of specific Trypanosoma cruzi trans-sialidase inhibitors.
Angew.Chem.Int.Ed.Engl., 47, 2008
|
|
3QQP
 
 | | Crystal Structure of 11beta-Hydroxysteroid Dehydrogenase 1 (11b-HSD1) in Complex with Urea Inhibitor | | Descriptor: | 3,4-dihydroquinolin-1(2H)-yl[4-(1H-imidazol-5-yl)piperidin-1-yl]methanone, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Loenze, P, Schimanski-Breves, S, Engel, C.K. | | Deposit date: | 2011-02-16 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Pyrrolidine-pyrazole ureas as potent and selective inhibitors of 11beta-hydroxysteroid-dehydrogenase type 1.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3D6Z
 
 | |
3CSI
 
 | |
2BGK
 
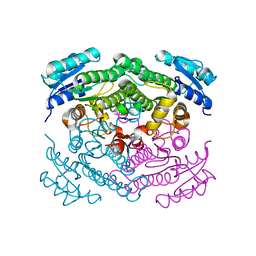 | | X-Ray structure of apo-Secoisolariciresinol Dehydrogenase | | Descriptor: | RHIZOME SECOISOLARICIRESINOL DEHYDROGENASE | | Authors: | Youn, B, Moinuddin, S.G, Davin, L.B, Lewis, N.G, Kang, C. | | Deposit date: | 2004-12-23 | | Release date: | 2005-01-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of Apo-Form and Binary/Ternary Complexes of Podophyllum Secoisolariciresinol Dehydrogenase, an Enzyme Involved in Formation of Health-Protecting and Plant Defense Lignans
J.Biol.Chem., 280, 2005
|
|
1RD9
 
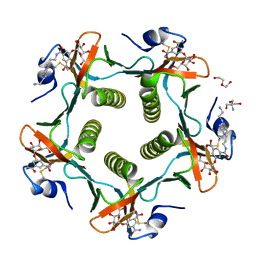 | | Cholera Toxin B-Pentamer Complexed With Bivalent Nitrophenol-Galactoside Ligand BV2 | | Descriptor: | 1,3-BIS-([3-(4-{3-[3-NITRO-5-(GALACTOPYRANOSYLOXY)-BENZOYLAMINO]-PROPYL}-PIPERAZIN-1-YL)-PROPYL-AMINO]-CARBONYLOXY)-2-AMINO-PROPANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, HEXAETHYLENE GLYCOL, ... | | Authors: | Pickens, J.C, Mitchell, D.D, Liu, J, Tan, X, Zhang, Z, Verlinde, C.L, Hol, W.G, Fan, E. | | Deposit date: | 2003-11-05 | | Release date: | 2004-10-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Nonspanning bivalent ligands as improved surface receptor binding inhibitors of the cholera toxin B pentamer.
Chem.Biol., 11, 2004
|
|
1RCV
 
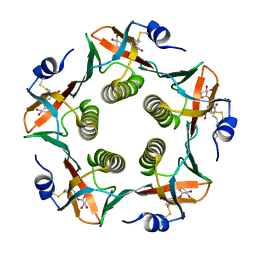 | | Cholera Toxin B-Pentamer Complexed With Bivalent Nitrophenol-Galactoside Ligand BV1 | | Descriptor: | [3-(4-{3-[3-NITRO-5-(GALACTOPYRANOSYLOXY)-BENZOYLAMINO]-PROPYL}-PIPERAZIN-1-YL)-PROPYLAMINO] -2-(3-{4-[3-(3-NITRO-5-[GALACTOPYRANOSYLOXY]-BENZOYLAMINO)-PROPYL]-PIPERAZIN-1-YL} -PROPYL-AMINO)-3,4-DIOXO-CYCLOBUTENE, cholera toxin B protein (CTB) | | Authors: | Pickens, J.C, Mitchell, D.D, Liu, J, Tan, X, Zhang, Z, Verlinde, C.L, Hol, W.G, Fan, E. | | Deposit date: | 2003-11-04 | | Release date: | 2004-10-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Nonspanning bivalent ligands as improved surface receptor binding inhibitors of the cholera toxin B pentamer.
Chem.Biol., 11, 2004
|
|
1S0I
 
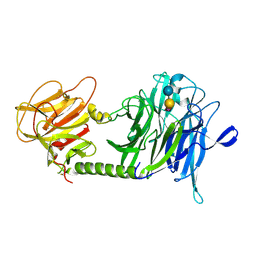 | | Trypanosoma cruzi trans-sialidase in complex with sialyl-lactose (Michaelis complex) | | Descriptor: | N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose, trans-sialidase | | Authors: | Amaya, M.F, Watts, A.G, Damager, I, Wehenkel, A, Nguyen, T, Buschiazzo, A, Paris, G, Frasch, A.C, Withers, S.G, Alzari, P.M. | | Deposit date: | 2003-12-31 | | Release date: | 2004-05-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Insights into the Catalytic Mechanism of Trypanosoma cruzi trans-Sialidase.
Structure, 12, 2004
|
|
2AH2
 
 | | Trypanosoma cruzi trans-sialidase in complex with 2,3-difluorosialic acid (covalent intermediate) | | Descriptor: | 5-acetamido-3,5-dideoxy-3-fluoro-D-erythro-alpha-L-manno-non-2-ulopyranosonic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Amaya, M.F, Watts, A.G, Damager, I, Wehenkel, A, Nguyen, T, Buschiazzo, A, Paris, G, Frasch, A.C, Withers, S.G, Alzari, P.M. | | Deposit date: | 2005-07-27 | | Release date: | 2005-08-23 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Insights into the Catalytic Mechanism of Trypanosoma cruzi trans-Sialidase
Structure, 12, 2004
|
|
3CSH
 
 | |
2BGM
 
 | | X-Ray structure of ternary-Secoisolariciresinol Dehydrogenase | | Descriptor: | MATAIRESINOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE (ACIDIC FORM), RHIZOME SECOISOLARICIRESINOL DEHYDROGENASE | | Authors: | Youn, B, Moinuddin, S.G, Davin, L.B, Lewis, N.G, Kang, C. | | Deposit date: | 2004-12-23 | | Release date: | 2005-01-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Apo-Form and Binary/Ternary Complexes of Podophyllum Secoisolariciresinol Dehydrogenase, an Enzyme Involved in Formation of Health-Protecting and Plant Defense Lignans
J.Biol.Chem., 280, 2005
|
|
3CUZ
 
 | |
3PJQ
 
 | | Trypanosoma cruzi trans-sialidase-like inactive isoform (including the natural mutation Tyr342His) in complex with lactose | | Descriptor: | Trans-sialidase, beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Oppezzo, P, Baraibar, M, Obal, G, Pritsch, O, Alzari, P.M, Buschiazzo, A. | | Deposit date: | 2010-11-10 | | Release date: | 2011-06-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of an enzymatically inactive trans-sialidase-like lectin from Trypanosoma cruzi: the carbohydrate binding mechanism involves residual sialidase activity.
Biochim.Biophys.Acta, 1814, 2011
|
|
3R6V
 
 | | Crystal structure of aspartase from Bacillus sp. YM55-1 with bound L-aspartate | | Descriptor: | ASPARTIC ACID, Aspartase, CALCIUM ION | | Authors: | Fibriansah, G, Puthan Veetil, V, Poelarends, G.J, Thunnissen, A.-M.W.H. | | Deposit date: | 2011-03-22 | | Release date: | 2011-07-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the catalytic mechanism of aspartate ammonia lyase.
Biochemistry, 50, 2011
|
|
3RLK
 
 | |
2QLI
 
 | | mPlum E16Q mutant | | Descriptor: | Fluorescent protein plum | | Authors: | Shu, X, Remington, S.J. | | Deposit date: | 2007-07-12 | | Release date: | 2008-07-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Structural Studies of Far-Red Emission in mPlum, a Monomeric Red Fluorescent Protein
To be Published
|
|
2BZS
 
 | | Binding of anti-cancer prodrug CB1954 to the activating enzyme NQO2 revealed by the crystal structure of their complex. | | Descriptor: | 5-(AZIRIDIN-1-YL)-2,4-DINITROBENZAMIDE, FLAVIN-ADENINE DINUCLEOTIDE, NRH DEHYDROGENASE [QUINONE] 2, ... | | Authors: | Abu Khader, M.M, Heap, J.T, De Matteis, C, Kellam, B, Doughty, S.W, Minton, N, Paoli, M. | | Deposit date: | 2005-08-22 | | Release date: | 2005-09-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding of the Anticancer Prodrug Cb1954 to the Activating Enzyme Nqo2 Revealed by the Crystal Structure of Their Complex.
J.Med.Chem., 48, 2005
|
|
1RDP
 
 | | Cholera Toxin B-Pentamer Complexed With Bivalent Nitrophenol-Galactoside Ligand BV3 | | Descriptor: | 1,3-BIS-([[3-(4-{3-[3-NITRO-5-(GALACTOPYRANOSYLOXY)-BENZOYLAMINO]-PROPYL}-PIPERAZIN-1-YL)-PROPYLAMINO-3,4-DIOXO-CYCLOBU TENYL]-AMINO-ETHYL]-AMINO-CARBONYLOXY)-2-AMINO-PROPANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, TRIETHYLENE GLYCOL, ... | | Authors: | Pickens, J.C, Mitchell, D.D, Liu, J, Tan, X, Zhang, Z, Verlinde, C.L, Hol, W.G, Fan, E. | | Deposit date: | 2003-11-05 | | Release date: | 2004-10-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Nonspanning bivalent ligands as improved surface receptor binding inhibitors of the cholera toxin B pentamer.
Chem.Biol., 11, 2004
|
|
3D71
 
 | | Crystal structure of E253Q BMRR bound to 22 base pair promoter site | | Descriptor: | BMR promoter DNA, CITRATE ANION, IMIDAZOLE, ... | | Authors: | Newberry, K.J, Huffman, J.L, Brennan, R.G. | | Deposit date: | 2008-05-20 | | Release date: | 2008-08-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of BmrR-Drug Complexes Reveal a Rigid Multidrug Binding Pocket and Transcription Activation through Tyrosine Expulsion
J.Biol.Chem., 283, 2008
|
|
3CV2
 
 | |
2B6N
 
 | | The 1.8 A crystal structure of a Proteinase K like enzyme from a psychrotroph Serratia species | | Descriptor: | CALCIUM ION, SULFATE ION, TRIPEPTIDE, ... | | Authors: | Helland, R, Larsen, A.N, Smalas, A.O, Willassen, N.P. | | Deposit date: | 2005-10-03 | | Release date: | 2006-03-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The 1.8 A crystal structure of a proteinase K-like enzyme from a psychrotroph Serratia species
Febs J., 273, 2006
|
|
2B9V
 
 | | Acetobacter turbidans alpha-amino acid ester hydrolase | | Descriptor: | Alpha-amino acid ester hydrolase | | Authors: | Barends, T.R.M. | | Deposit date: | 2005-10-13 | | Release date: | 2005-12-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Acetobacter turbidans alpha-amino acid ester hydrolase: how a single mutation improves an antibiotic-producing enzyme.
J.Biol.Chem., 281, 2006
|
|
1SH7
 
 | |
3O73
 
 | | Crystal structure of quinone reductase 2 in complex with the indolequinone MAC627 | | Descriptor: | 5-[(4-aminobutyl)amino]-1,2-dimethyl-3-[(4-nitrophenoxy)methyl]-1H-indole-4,7-dione, FLAVIN-ADENINE DINUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Dufour, M, Yan, C, Colucci, M.A, Siegel, D, Li, Y, De Matteis, C.I, Ross, D, Moody, C.J. | | Deposit date: | 2010-07-30 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism-Based Inhibition of Quinone Reductase 2 (NQO2): Selectivity for NQO2 over NQO1 and Structural Basis for Flavoprotein Inhibition.
Chembiochem, 12, 2011
|
|
2VFT
 
 | |
