6XJP
 
 | |
6XJR
 
 | |
6XJU
 
 | | Crystal Structure of KPT-8602 bound to CRM1 (E582K, 537-DLTVK-541 to GLCEQ) | | Descriptor: | (2R)-3-{3-[3,5-bis(trifluoromethyl)phenyl]-1H-1,2,4-triazol-1-yl}-2-(pyrimidin-5-yl)propanamide, Exportin-1, GTP-binding nuclear protein Ran, ... | | Authors: | Baumhardt, J.M, Chook, Y.M. | | Deposit date: | 2020-06-24 | | Release date: | 2021-01-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.193 Å) | | Cite: | Recurrent XPO1 mutations alter pathogenesis of chronic lymphocytic leukemia.
J Hematol Oncol, 14, 2021
|
|
6XJS
 
 | |
4UJ5
 
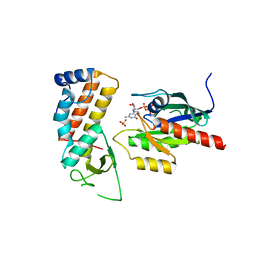 | | Crystal structure of human Rab11-Rabin8-FIP3 | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, RAB-3A-INTERACTING PROTEIN, ... | | Authors: | Vetter, M, Lorentzen, E. | | Deposit date: | 2015-04-08 | | Release date: | 2015-08-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Structure of Rab11-Fip3-Rabin8 Reveals Simultaneous Binding of Fip3 and Rabin8 Effectors to Rab11.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4UJ4
 
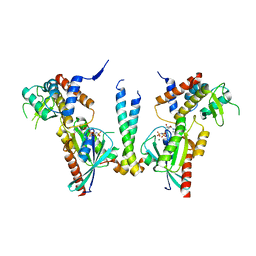 | | Crystal structure of human Rab11-Rabin8-FIP3 | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Rab-3A-interacting protein, ... | | Authors: | Vetter, M, Lorentzen, E. | | Deposit date: | 2015-04-08 | | Release date: | 2015-08-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Structure of Rab11-FIP3-Rabin8 reveals simultaneous binding of FIP3 and Rabin8 effectors to Rab11.
Nat. Struct. Mol. Biol., 22, 2015
|
|
4UJ3
 
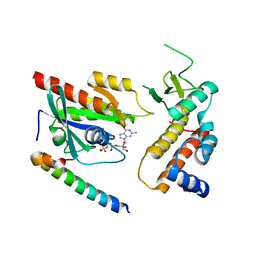 | | Crystal structure of human Rab11-Rabin8-FIP3 | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, RAB-3A-INTERACTING PROTEIN, ... | | Authors: | Vetter, M, Lorentzen, E. | | Deposit date: | 2015-04-08 | | Release date: | 2015-08-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of Rab11-Fip3-Rabin8 Reveals Simultaneous Binding of Fip3 and Rabin8 Effectors to Rab11.
Nat.Struct.Mol.Biol., 22, 2015
|
|
1YZK
 
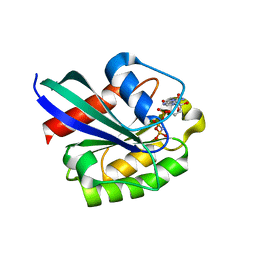 | | GppNHp bound Rab11 GTPase | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related protein Rab-11A | | Authors: | Eathiraj, S, Pan, X, Ritacco, C, Lambright, D.G. | | Deposit date: | 2005-02-28 | | Release date: | 2005-07-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of family-wide Rab GTPase recognition by rabenosyn-5.
Nature, 436, 2005
|
|
6CPN
 
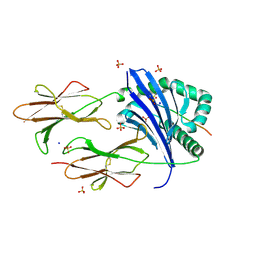 | | Crystal structure of DR11 presenting the RQ13 peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Gag polyprotein, ... | | Authors: | Farenc, C, Gras, S, Rossjohn, J. | | Deposit date: | 2018-03-13 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CD4+T cell-mediated HLA class II cross-restriction in HIV controllers.
Sci Immunol, 3, 2018
|
|
6CQN
 
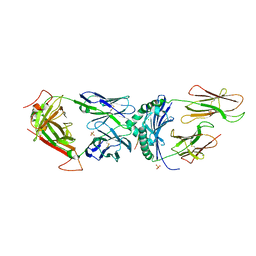 | | Crystal structure of F5 TCR -DR11-RQ13 peptide complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, F5 alpha chain, ... | | Authors: | Farenc, C, Gras, S, Rossjohn, J. | | Deposit date: | 2018-03-15 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | CD4+T cell-mediated HLA class II cross-restriction in HIV controllers.
Sci Immunol, 3, 2018
|
|
1SJE
 
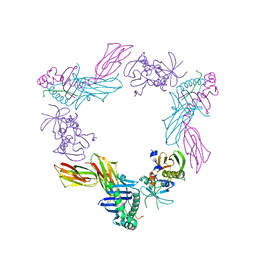 | | HLA-DR1 complexed with a 16 residue HIV capsid peptide bound in a hairpin conformation | | Descriptor: | Enterotoxin type C-3, GAG polyprotein, HLA class II histocompatibility antigen, ... | | Authors: | Zavala-Ruiz, Z, Strug, I, Walker, B.D, Norris, P.J, Stern, L.J. | | Deposit date: | 2004-03-03 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A hairpin turn in a class II MHC-bound peptide orients residues outside the binding groove for T cell recognition.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
6CPL
 
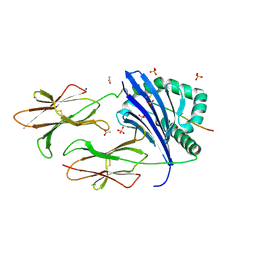 | | Crystal structure of DR11 presenting the gag293 epitope | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Farenc, C, Gras, S, Rossjohn, J. | | Deposit date: | 2018-03-13 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | CD4+T cell-mediated HLA class II cross-restriction in HIV controllers.
Sci Immunol, 3, 2018
|
|
6CQL
 
 | | Crystal structure of F24 TCR -DR11-RQ13 peptide complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, F24 alpha chain, F24 beta Chain, ... | | Authors: | Farenc, C, Gras, S, Rossjohn, J. | | Deposit date: | 2018-03-15 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CD4+T cell-mediated HLA class II cross-restriction in HIV controllers.
Sci Immunol, 3, 2018
|
|
6CQJ
 
 | | Crystal structure of DR1 presenting the RQ13 peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Farenc, C, Gras, S, Rossjohn, J. | | Deposit date: | 2018-03-15 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | CD4+T cell-mediated HLA class II cross-restriction in HIV controllers.
Sci Immunol, 3, 2018
|
|
8PR1
 
 | | Cytoplasmic dynein-B heavy chain bound to IC-LC tower | | Descriptor: | Cytoplasmic dynein 1 heavy chain 1, Cytoplasmic dynein 1 intermediate chain 2, Cytoplasmic dynein 1 light intermediate chain 2, ... | | Authors: | Singh, K, Lau, C.K, Manigrasso, G, Gassmann, R, Carter, A.P. | | Deposit date: | 2023-07-12 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | Molecular mechanism of dynein-dynactin complex assembly by LIS1.
Science, 383, 2024
|
|
6CQR
 
 | | Crystal structure of F24 TCR -DR1-RQ13 peptide complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, F24 alpha chain, F24 beta chain, ... | | Authors: | Farenc, C, Gras, S, Rossjohn, J. | | Deposit date: | 2018-03-16 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | CD4+T cell-mediated HLA class II cross-restriction in HIV controllers.
Sci Immunol, 3, 2018
|
|
5DHF
 
 | |
1R5I
 
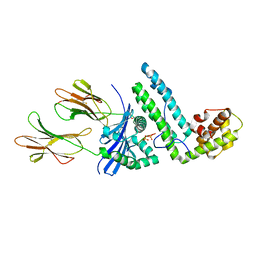 | | Crystal structure of the MAM-MHC complex | | Descriptor: | HLA class II histocompatibility antigen, DR alpha chain, DRB1-1 beta chain, ... | | Authors: | Zhao, Y, Li, Z, Drozd, S.J, Guo, Y, Mourad, W, Li, H. | | Deposit date: | 2003-10-10 | | Release date: | 2004-03-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Mycoplasma arthritidis mitogen complexed with HLA-DR1 reveals a novel superantigen fold and a dimerized superantigen-MHC complex.
Structure, 12, 2004
|
|
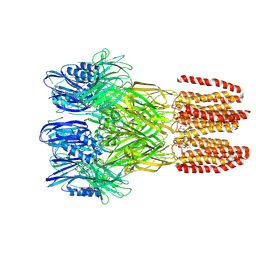
6V4A
 
 | |
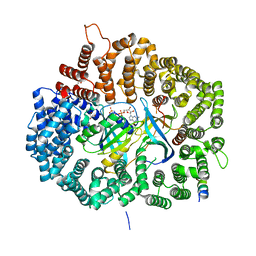
5DHA
 
 | |
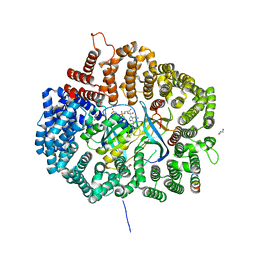
5DIF
 
 | |
5DI9
 
 | |
8RGG
 
 | | Structure of dynein-2 intermediate chain DYNC2I2 (WDR34) in complex with dynein-2 heavy chain DYNC2H1. | | Descriptor: | Cytoplasmic dynein 2 intermediate chain 1, Cytoplasmic dynein 2 intermediate chain 2, Dynein light chain 1, ... | | Authors: | Mukhopadhyay, A.G, Toropova, K, Daly, L, Wells, J, Vuolo, L, Mladenov, M, Seda, M, Jenkins, D, Stephens, D.J, Roberts, A.J. | | Deposit date: | 2023-12-13 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure and tethering mechanism of dynein-2 intermediate chains in intraflagellar transport.
Embo J., 43, 2024
|
|
8RGH
 
 | | Structure of dynein-2 intermediate chain DYNC2I1 (WDR60) in complex with the dynein-2 heavy chain DYNC2H1. | | Descriptor: | Cytoplasmic dynein 2 intermediate chain 1, Cytoplasmic dynein 2 intermediate chain 2, Cytoplasmic dynein 2 light intermediate chain 1, ... | | Authors: | Mukhopadhyay, A.G, Toropova, K, Daly, L, Wells, J, Vuolo, L, Seda, M, Jenkins, D, Stephens, D.J, Roberts, A.J. | | Deposit date: | 2023-12-13 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure and tethering mechanism of dynein-2 intermediate chains in intraflagellar transport.
Embo J., 43, 2024
|
|
8PTK
 
 | | Composite structure of Dynein-Dynactin-JIP3-LIS1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ARP1 actin related protein 1 homolog A, ... | | Authors: | Singh, K, Lau, C.K, Manigrasso, G, Gassmann, R, Carter, A.P. | | Deposit date: | 2023-07-14 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Molecular mechanism of dynein-dynactin complex assembly by LIS1.
Science, 383, 2024
|
|
