6SRZ
 
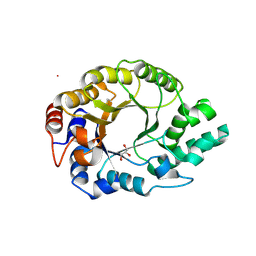 | | Kemp Eliminase HG3.17 mutant Q50H, E47N, N300D Complexed with Transition State Analog 6-Nitrobenzotriazole | | Descriptor: | 6-NITROBENZOTRIAZOLE, GLYCEROL, Kemp Eliminase HG3.17 Q50H, ... | | Authors: | Bloch, J.S, Pinkas, D.M, Hilvert, D. | | Deposit date: | 2019-09-06 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Contribution of Oxyanion Stabilization to Kemp Eliminase Efficiencyproficiency
Acs Catalysis, 2020
|
|
1N0A
 
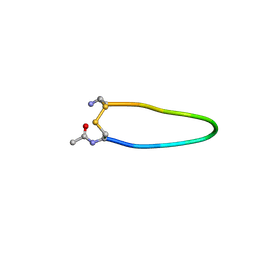 | |
4O60
 
 | | Structure of ankyrin repeat protein | | Descriptor: | ANK-N5C-317 | | Authors: | Ethayathulla, A.S, Guan, L. | | Deposit date: | 2013-12-20 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | A transcription blocker isolated from a designed repeat protein combinatorial library by in vivo functional screen.
Sci Rep, 5, 2015
|
|
4O5S
 
 | |
4LPU
 
 | |
4LPY
 
 | |
5TWI
 
 | |
5U5B
 
 | | Coiled Coil Peptide Metal Coordination Framework: Trimer Fold | | Descriptor: | COPPER (II) ION, Designed trimeric coiled coil peptide with two terpyridine side chains, GLYCEROL | | Authors: | Tavenor, N.A, Murnin, M.J, Horne, W.S. | | Deposit date: | 2016-12-06 | | Release date: | 2017-02-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Supramolecular Metal-Coordination Polymers, Nets, and Frameworks from Synthetic Coiled-Coil Peptides.
J. Am. Chem. Soc., 139, 2017
|
|
4LPV
 
 | | Crystal structure of TENCON variant P41BR3-42 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, TENCON variant P41BR3-42 | | Authors: | Teplyakov, A, Obmolova, G, Luo, J, Gilliland, G.L. | | Deposit date: | 2013-07-16 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | C-terminal beta-strand swapping in a consensus-derived fibronectin Type III scaffold.
Proteins, 82, 2014
|
|
5US3
 
 | |
5VAV
 
 | |
4OXW
 
 | |
6U47
 
 | |
4OYD
 
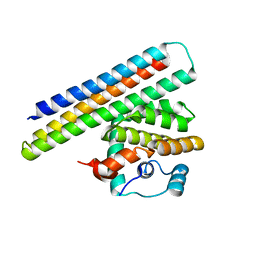 | | Crystal structure of a computationally designed inhibitor of an Epstein-Barr viral Bcl-2 protein | | Descriptor: | 1,2-ETHANEDIOL, Apoptosis regulator BHRF1, Computationally designed Inhibitor | | Authors: | Shen, B, Procko, E, Baker, D, Stoddard, B. | | Deposit date: | 2014-02-11 | | Release date: | 2014-07-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A computationally designed inhibitor of an epstein-barr viral bcl-2 protein induces apoptosis in infected cells.
Cell, 157, 2014
|
|
5V65
 
 | |
5VNW
 
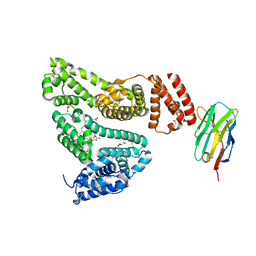 | |
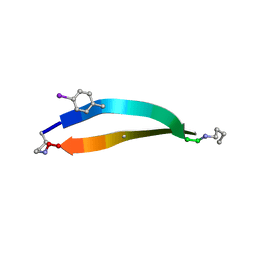
3CAY
 
 | | Crystal structure of Lipopeptide Detergent (LPD-12) | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, LPD-12 | | Authors: | Ho, D.N, Pomroy, N.C, Cuesta-Seijo, J.A, Prive, G.G. | | Deposit date: | 2008-02-20 | | Release date: | 2008-09-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of a self-assembling lipopeptide detergent at 1.20 A.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
4OZC
 
 | | Backbone Modifications in the Protein GB1 Helix and Loops: beta-ACPC21, beta-ACPC24, beta-3-Lys28, beta-3-Lys31, beta-ACPC35, beta-ACPC40 | | Descriptor: | GLYCEROL, SULFATE ION, Streptococcal Protein GB1 Backbone Modified Variant: beta-ACPC21, ... | | Authors: | Reinert, Z.E, Horne, W.S. | | Deposit date: | 2014-02-14 | | Release date: | 2014-07-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Folding Thermodynamics of Protein-Like Oligomers with Heterogeneous Backbones.
Chem Sci, 5, 2014
|
|
3CBA
 
 | |
6A9W
 
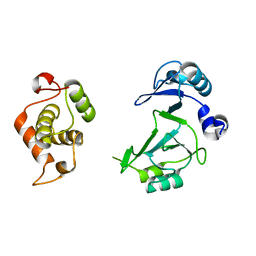 | | Structure of the bifunctional DNA primase-polymerase from phage NrS-1 | | Descriptor: | Primase | | Authors: | Guo, H.J, Li, M.J, Wang, T.L, Wu, H, Zhou, H, Xu, C.Y, Liu, X.P, Yu, F, He, J.H. | | Deposit date: | 2018-07-16 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure and biochemical studies of the bifunctional DNA primase-polymerase from phage NrS-1.
Biochem. Biophys. Res. Commun., 510, 2019
|
|
4IWW
 
 | | Computational Design of an Unnatural Amino Acid Metalloprotein with Atomic Level Accuracy | | Descriptor: | COBALT (II) ION, SULFATE ION, Unnatural Amino Acid Mediated Metalloprotein | | Authors: | Mills, J, Bolduc, J, Khare, S, Stoddard, B, Baker, D. | | Deposit date: | 2013-01-24 | | Release date: | 2013-08-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Computational design of an unnatural amino Acid dependent metalloprotein with atomic level accuracy.
J.Am.Chem.Soc., 135, 2013
|
|
4OZA
 
 | | Backbone Modifications in the Protein GB1 Helix: beta-3-Ala24, beta-3-Lys28, beta-3-Gln32, beta-3-Asp36 | | Descriptor: | ISOPROPYL ALCOHOL, Streptococcal Protein GB1 Backbone Modified Variant: beta-3-Ala24, beta-3-Lys28, ... | | Authors: | Reinert, Z.E, Horne, W.S. | | Deposit date: | 2014-02-14 | | Release date: | 2014-07-16 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Folding Thermodynamics of Protein-Like Oligomers with Heterogeneous Backbones.
Chem Sci, 5, 2014
|
|
5VNV
 
 | | Crystal structure of Nb.b201 | | Descriptor: | FORMIC ACID, Nb.b201, PENTAETHYLENE GLYCOL, ... | | Authors: | Kruse, A.C, McMahon, C. | | Deposit date: | 2017-05-01 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Yeast surface display platform for rapid discovery of conformationally selective nanobodies.
Nat. Struct. Mol. Biol., 25, 2018
|
|
4NW9
 
 | |
4P4X
 
 | |
