5N1O
 
 | |
9C6T
 
 | |
5N1K
 
 | |
4DZK
 
 | | A de novo designed Coiled Coil CC-Tri-N13 | | Descriptor: | CHLORIDE ION, COILED-COIL PEPTIDE CC-TRI-N13 | | Authors: | Bruning, M, Thomson, A.R, Zaccai, N.R, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2012-03-01 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | A basis set of de novo coiled-coil Peptide oligomers for rational protein design and synthetic biology.
ACS Synth Biol, 1, 2012
|
|
4WKG
 
 | |
6Q78
 
 | |
8W11
 
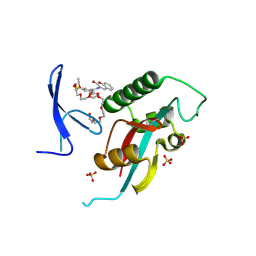 | | Structure of human PIN1 covalently derivatized with SuFEx compound | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 3-(fluorosulfonyl)-5-[(2-oxo-2H-1-benzopyran-3-yl)carbamoyl]benzoic acid, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, ... | | Authors: | Louie, G.V, Noel, J.P, Wang, W.-M, Kelly, J.W. | | Deposit date: | 2024-02-14 | | Release date: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of human PIN1 covalently derivatized with SuFEx compound
To be published
|
|
9FVF
 
 | | UMG-SP3 amidase from uncultured bacterium | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, Amidase UMG-SP3, SULFATE ION | | Authors: | Rotilio, L, Morth, J.P. | | Deposit date: | 2024-06-27 | | Release date: | 2025-02-05 | | Last modified: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural and Functional Characterization of an Amidase Targeting a Polyurethane for Sustainable Recycling.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
6QG4
 
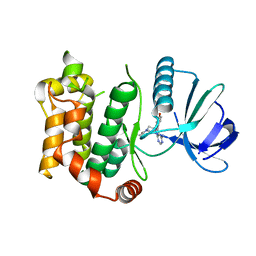 | |
6NTK
 
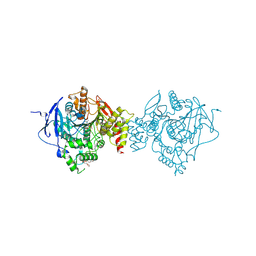 | | Crystal Structure of Recombinant Human Acetylcholinesterase Inhibited by A-232 | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bester, S.M, Guelta, M.A, Height, J.J, Pegan, S.D. | | Deposit date: | 2019-01-29 | | Release date: | 2020-07-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.406 Å) | | Cite: | Insights into inhibition of human acetylcholinesterase by Novichok, A-series Nerve Agents
To Be Published
|
|
6ABH
 
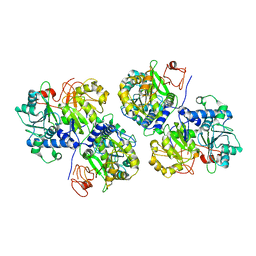 | |
6NWT
 
 | | RORgamma Ligand Binding Domain | | Descriptor: | 1,1,1,3,3,3-hexafluoro-2-[2-fluoro-4'-({4-[(pyridin-4-yl)methyl]piperazin-1-yl}methyl)[1,1'-biphenyl]-4-yl]propan-2-ol, Nuclear receptor ROR-gamma | | Authors: | Strutzenberg, T.S, Park, H, Griffin, P.R. | | Deposit date: | 2019-02-07 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | HDX-MS reveals structural determinants for ROR gamma hyperactivation by synthetic agonists.
Elife, 8, 2019
|
|
6YJZ
 
 | |
6HMR
 
 | | Crystal structure of human Casein Kinase I delta in complex with a photoswitchable 2-Azothiazole-based inhibitor (compound 2) | | Descriptor: | 3-(2,5-dimethoxyphenyl)-~{N}-[4-[4-(4-fluorophenyl)-2-[(~{E})-phenyldiazenyl]-1,3-thiazol-5-yl]pyridin-2-yl]propanamide, Casein kinase I isoform delta, MALONIC ACID | | Authors: | Pichlo, C, Schehr, M, Charl, J, Brunstein, E, Peifer, C, Baumann, U. | | Deposit date: | 2018-09-12 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.782 Å) | | Cite: | 2-Azo-, 2-diazocine-thiazols and 2-azo-imidazoles as photoswitchable kinase inhibitors: limitations and pitfalls of the photoswitchable inhibitor approach.
Photochem. Photobiol. Sci., 18, 2019
|
|
5N66
 
 | | Crystal Structure of p38alpha in Complex with Lipid Pocket Ligand 9j | | Descriptor: | 1-(5-TERT-BUTYL-2-P-TOLYL-2H-PYRAZOL-3-YL)-3-[4-(2-MORPHOLIN-4-YL-ETHOXY)-NAPHTHALEN-1-YL]-UREA, Mitogen-activated protein kinase 14, ~{N}4-[[4-(cyclopropylmethyl)furan-2-yl]methyl]-2-phenyl-quinazoline-4,7-diamine | | Authors: | Buehrmann, M, Mueller, M.P, Wiedemann, B, Rauh, D. | | Deposit date: | 2017-02-14 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based design, synthesis and crystallization of 2-arylquinazolines as lipid pocket ligands of p38 alpha MAPK.
PLoS ONE, 12, 2017
|
|
7PGN
 
 | | HHP-C in complex with glycosaminoglycan mimic SOS | | Descriptor: | 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Griffiths, S.C, Schwab, R.A, El Omari, K, Bishop, B, Iverson, E.J, Malinuskas, T, Dubey, R, Qian, M, Covey, D.F, Gilbert, R.J.C, Rohatgi, R, Siebold, C. | | Deposit date: | 2021-08-14 | | Release date: | 2021-12-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Hedgehog-Interacting Protein is a multimodal antagonist of Hedgehog signalling.
Nat Commun, 12, 2021
|
|
6O0P
 
 | | crystal structure of BCL-2 G101A mutation with venetoclax | | Descriptor: | 4-{4-[(4'-chloro-5,5-dimethyl[3,4,5,6-tetrahydro[1,1'-biphenyl]]-2-yl)methyl]piperazin-1-yl}-N-[(3-nitro-4-{[(oxan-4-yl )methyl]amino}phenyl)sulfonyl]-2-[(1H-pyrrolo[2,3-b]pyridin-5-yl)oxy]benzamide, Apoptosis regulator Bcl-2,Bcl-2-like protein 1,Apoptosis regulator Bcl-2, DI(HYDROXYETHYL)ETHER | | Authors: | Birkinshaw, R.W, Luo, C.S, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2019-02-17 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of BCL-2 in complex with venetoclax reveal the molecular basis of resistance mutations.
Nat Commun, 10, 2019
|
|
6H26
 
 | | Rabbit muscle phosphoglycerate mutase | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Phosphoglycerate mutase | | Authors: | Wisniewski, J, Barciszewski, J, Jaskolski, M, Rakus, D. | | Deposit date: | 2018-07-13 | | Release date: | 2019-07-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.288 Å) | | Cite: | Rabbit muscle phosphoglycerate mutase
To Be Published
|
|
9LID
 
 | |
6O0K
 
 | | crystal structure of BCL-2 with venetoclax | | Descriptor: | 4-{4-[(4'-chloro-5,5-dimethyl[3,4,5,6-tetrahydro[1,1'-biphenyl]]-2-yl)methyl]piperazin-1-yl}-N-[(3-nitro-4-{[(oxan-4-yl )methyl]amino}phenyl)sulfonyl]-2-[(1H-pyrrolo[2,3-b]pyridin-5-yl)oxy]benzamide, Apoptosis regulator Bcl-2, NONAETHYLENE GLYCOL | | Authors: | Birkinshaw, R.W, Luo, C.S, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2019-02-16 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structures of BCL-2 in complex with venetoclax reveal the molecular basis of resistance mutations.
Nat Commun, 10, 2019
|
|
6Z72
 
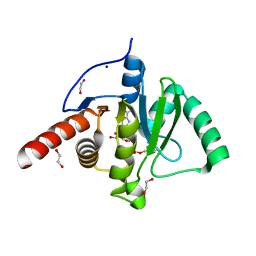 | | SARS-CoV-2 Macrodomain in complex with ADP-HPM | | Descriptor: | 1,2-ETHANEDIOL, Adenosine Diphosphate (Hydroxymethyl)pyrrolidine monoalcohol, D-MALATE, ... | | Authors: | Zorzini, V, Rack, J, Ahel, I. | | Deposit date: | 2020-05-29 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Viral macrodomains: a structural and evolutionary assessment of the pharmacological potential.
Open Biology, 10, 2020
|
|
5NK0
 
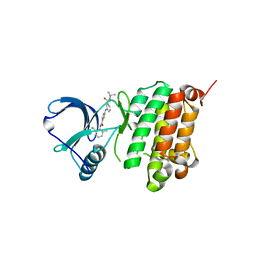 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 1j | | Descriptor: | 2-[[3-[(3-azanyl-2,2-dimethyl-propyl)carbamoyl]phenyl]amino]-~{N}-(2-chloranyl-6-methyl-phenyl)-1,3-thiazole-5-carboxamide, Ephrin type-A receptor 2 | | Authors: | Kudlinzki, D, Linhard, V.L, Witt, K, Gande, S.L, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.597 Å) | | Cite: | Chemoproteomics-Aided Medicinal Chemistry for the Discovery of EPHA2 Inhibitors.
ChemMedChem, 12, 2017
|
|
7RB6
 
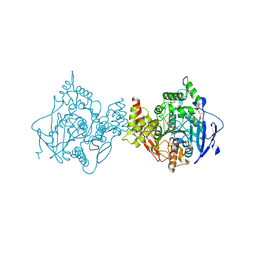 | | Low temperature structure of hAChE in complex with substrate analog 4K-TMA | | Descriptor: | 4,4-DIHYDROXY-N,N,N-TRIMETHYLPENTAN-1-AMINIUM, Acetylcholinesterase, GLYCEROL, ... | | Authors: | Kovalevsky, A, Gerlits, O, Radic, Z. | | Deposit date: | 2021-07-05 | | Release date: | 2021-09-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Room temperature crystallography of human acetylcholinesterase bound to a substrate analogue 4K-TMA: Towards a neutron structure
Curr Res Struct Biol, 3, 2021
|
|
6O66
 
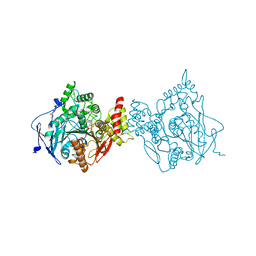 | |
6HD1
 
 | | human STEAP4 bound to NADPH, FAD and heme. | | Descriptor: | (2R)-3-(phosphonooxy)propane-1,2-diyl dihexanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Oosterheert, W, van Bezouwen, L.S, Rodenburg, R.N.P, Forster, F, Mattevi, A, Gros, P. | | Deposit date: | 2018-08-17 | | Release date: | 2018-10-24 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures of human STEAP4 reveal mechanism of iron(III) reduction.
Nat Commun, 9, 2018
|
|
