5OB3
 
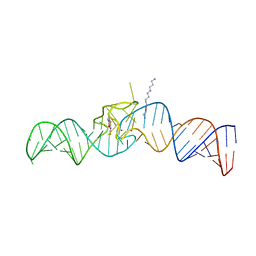 | | iSpinach aptamer | | Descriptor: | 4-(3,5-difluoro-4-hydroxybenzyl)-1,2-dimethyl-1H-imidazol-5-ol, POTASSIUM ION, RNA aptamer (69-MER), ... | | Authors: | Fernandez-Millan, P, Autour, A, Westhof, E, Ryckelynck, M. | | Deposit date: | 2017-06-26 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | Crystal structure and fluorescence properties of the iSpinach aptamer in complex with DFHBI.
RNA, 23, 2017
|
|
4MYV
 
 | | Free HSV-2 gD structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein D | | Authors: | Lu, G, Zhang, N, Qi, J, Li, Y, Chen, Z, Zheng, C, Yan, J, Gao, G.F. | | Deposit date: | 2013-09-28 | | Release date: | 2014-10-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Crystal structure of herpes simplex virus 2 gD bound to nectin-1 reveals a conserved mode of receptor recognition.
J.Virol., 88, 2014
|
|
5OBB
 
 | |
2Z5Q
 
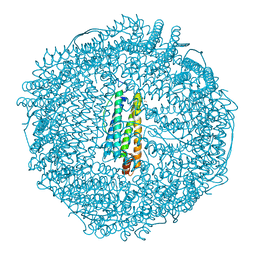 | | Apo-Fr with intermediate content of Pd ion | | Descriptor: | CADMIUM ION, Ferritin light chain, GLYCEROL, ... | | Authors: | Ueno, T, Hirata, K, Abe, M, Suzuki, M, Abe, S, Shimizu, N, Yamamoto, M, Takata, M, Watanabe, Y. | | Deposit date: | 2007-07-16 | | Release date: | 2008-07-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Process of accumulation of metal ions on the interior surface of apo-ferritin: crystal structures of a series of apo-ferritins containing variable quantities of Pd(II) ions.
J.Am.Chem.Soc., 131, 2009
|
|
8TPJ
 
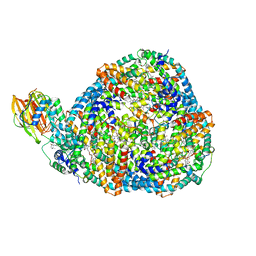 | | Top cylinder bound to OCP from high-resolution phycobilisome quenched by OCP (local refinement) | | Descriptor: | Allophycocyanin alpha chain, Allophycocyanin beta chain, Orange carotenoid-binding protein, ... | | Authors: | Sauer, P.V, Sutter, M, Cupellini, L. | | Deposit date: | 2023-08-04 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Structural and quantum chemical basis for OCP-mediated quenching of phycobilisomes.
Sci Adv, 10, 2024
|
|
4H5Q
 
 | |
1PTJ
 
 | | Crystal structure analysis of the DI and DIII complex of transhydrogenase with a thio-nicotinamide nucleotide analogue | | Descriptor: | GLYCEROL, NAD(P) transhydrogenase subunit alpha part 1, NAD(P) transhydrogenase subunit beta, ... | | Authors: | Singh, A, Venning, J.D, Quirk, P.G, van Boxel, G.I, Rodrigues, D.J, White, S.A, Jackson, J.B. | | Deposit date: | 2003-06-23 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Interactions between transhydrogenase and thio-nicotinamide analogues of NAD(H) and NADP(H) underline the importance of nucleotide conformational changes in coupling to proton translocation
J.Biol.Chem., 278, 2003
|
|
4H80
 
 | |
5AC8
 
 | | S. enterica HisA with mutations D10G, dup13-15, G102A | | Descriptor: | 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase, SULFATE ION | | Authors: | Newton, M, Guo, X, Soderholm, A, Nasvall, J, Andersson, D, Patrick, W, Selmer, M. | | Deposit date: | 2015-08-12 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Structural and functional innovations in the real-time evolution of new ( beta alpha )8 barrel enzymes.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5ODH
 
 | | Heterodisulfide reductase / [NiFe]-hydrogenase complex from Methanothermococcus thermolithotrophicus soaked with heterodisulfide for 3.5 minutes | | Descriptor: | 1-THIOETHANESULFONIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, ... | | Authors: | Wagner, T, Koch, J, Ermler, U, Shima, S. | | Deposit date: | 2017-07-05 | | Release date: | 2017-08-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Methanogenic heterodisulfide reductase (HdrABC-MvhAGD) uses two noncubane [4Fe-4S] clusters for reduction.
Science, 357, 2017
|
|
8TQD
 
 | | NF-Kappa-B1 Bound with a Covalent Inhibitor | | Descriptor: | 1-(2-bromo-4-chlorophenyl)-N-{(3S)-1-[(E)-iminomethyl]pyrrolidin-3-yl}methanesulfonamide, Nuclear factor NF-kappa-B p105 subunit | | Authors: | Hilbert, B.J. | | Deposit date: | 2023-08-07 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | DrugMap: A quantitative pan-cancer analysis of cysteine ligandability.
Cell, 187, 2024
|
|
2Z75
 
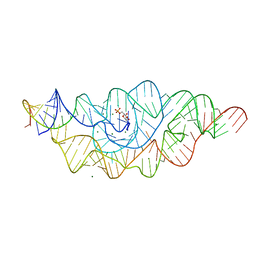 | | T. tengcongensis glmS ribozyme bound to glucosamine-6-phosphate | | Descriptor: | 2-amino-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, MAGNESIUM ION, glmS ribozyme RNA, ... | | Authors: | Klein, D.J, Wilkinson, S.R, Been, M.D, Ferre-D'Amare, A.R. | | Deposit date: | 2007-08-15 | | Release date: | 2007-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Requirement of helix P2.2 and nucleotide G1 for positioning the cleavage site and cofactor of the glmS ribozyme
J.Mol.Biol., 373, 2007
|
|
8TFM
 
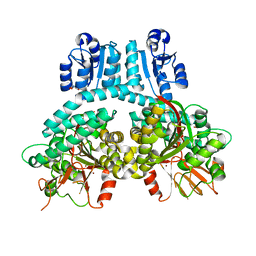 | | PorX with Zn (primitive orthorhombic crystal form) | | Descriptor: | GLYCEROL, Response regulator receiver protein, ZINC ION | | Authors: | Saran, A, Zeytuni, N. | | Deposit date: | 2023-07-11 | | Release date: | 2024-04-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Unveiling the molecular mechanisms of the type IX secretion system's response regulator: Structural and functional insights.
Pnas Nexus, 3, 2024
|
|
5O0J
 
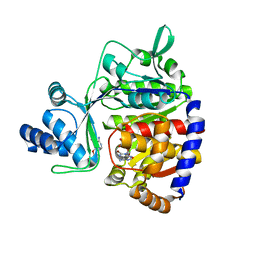 | | ADP-dependent glucokinase from Pyrococcus horikoshii | | Descriptor: | 8-BROMO-ADENOSINE-5'-MONOPHOSPHATE, ADP-dependent glucokinase, alpha-D-glucopyranose | | Authors: | Grudnik, P, Dubin, G. | | Deposit date: | 2017-05-16 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural basis for ADP-dependent glucokinase inhibition by 8-bromo-substituted adenosine nucleotide.
J. Biol. Chem., 293, 2018
|
|
2ZHX
 
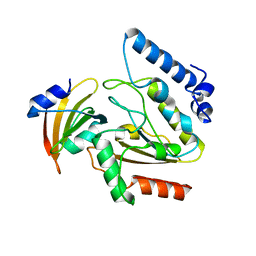 | | Crystal structure of Uracil-DNA Glycosylase from Mycobacterium tuberculosis in complex with a proteinaceous inhibitor | | Descriptor: | Uracil-DNA glycosylase, Uracil-DNA glycosylase inhibitor | | Authors: | Kaushal, P.S, Talawar, R.K, Krishna, P.D.V, Varshney, U, Vijayan, M. | | Deposit date: | 2008-02-11 | | Release date: | 2008-05-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Unique features of the structure and interactions of mycobacterial uracil-DNA glycosylase: structure of a complex of the Mycobacterium tuberculosis enzyme in comparison with those from other sources
Acta Crystallogr.,Sect.D, 64, 2008
|
|
8TED
 
 | | PorX primitive orthorhombic crystal form | | Descriptor: | ACETATE ION, BROMIDE ION, CALCIUM ION, ... | | Authors: | Saran, A, Zeytuni, N. | | Deposit date: | 2023-07-06 | | Release date: | 2024-04-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Unveiling the molecular mechanisms of the type IX secretion system's response regulator: Structural and functional insights.
Pnas Nexus, 3, 2024
|
|
3VDB
 
 | | E. coli (lacZ) beta-galactosidase (N460T) in complex with galactonolactone | | Descriptor: | Beta-galactosidase, D-galactonolactone, DIMETHYL SULFOXIDE, ... | | Authors: | Wheatley, R.W, Kappelhoff, J.C, Hahn, J.N, Dugdale, M.L, Dutkoski, M.J, Tamman, S.D, Fraser, M.E, Huber, R.E. | | Deposit date: | 2012-01-04 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Substitution for Asn460 cripples {beta}-galactosidase (Escherichia coli) by increasing substrate affinity and decreasing transition state stability.
Arch.Biochem.Biophys., 521, 2012
|
|
5O0Q
 
 | |
3VD7
 
 | | E. coli (lacZ) beta-galactosidase (N460S) in complex with galactotetrazole | | Descriptor: | (5R, 6S, 7S, ... | | Authors: | Wheatley, R.W, Kappelhoff, J.C, Hahn, J.N, Dugdale, M.L, Dutkoski, M.J, Tamman, S.D, Fraser, M.E, Huber, R.E. | | Deposit date: | 2012-01-04 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Substitution for Asn460 cripples {beta}-galactosidase (Escherichia coli) by increasing substrate affinity and decreasing transition state stability.
Arch.Biochem.Biophys., 521, 2012
|
|
4HH1
 
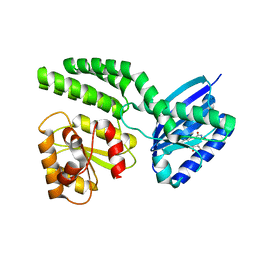 | | Dark-state structure of AppA wild-type without the Cys-rich region from Rb. sphaeroides | | Descriptor: | AppA protein, FLAVIN MONONUCLEOTIDE | | Authors: | Winkler, A, Heintz, U, Lindner, R, Reinstein, J, Shoeman, R, Schlichting, I. | | Deposit date: | 2012-10-09 | | Release date: | 2013-06-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.501 Å) | | Cite: | A ternary AppA-PpsR-DNA complex mediates light regulation of photosynthesis-related gene expression.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4GPT
 
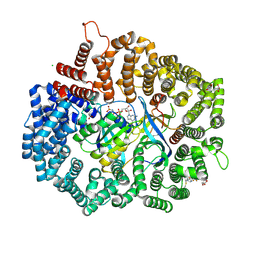 | | Crystal structure of KPT251 in complex with CRM1-Ran-RanBP1 | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-{3-[3,5-bis(trifluoromethyl)phenyl]-1H-1,2,4-triazol-1-yl}ethyl)-1,3,4-oxadiazole, CHLORIDE ION, ... | | Authors: | Sun, Q, Chook, Y.M. | | Deposit date: | 2012-08-21 | | Release date: | 2012-09-05 | | Last modified: | 2013-01-23 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Antileukemic activity of nuclear export inhibitors that spare normal hematopoietic cells.
Leukemia, 27, 2013
|
|
4HAI
 
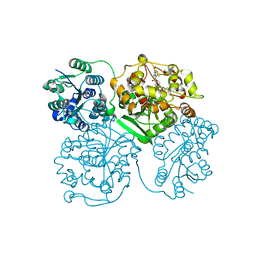 | | Crystal structure of human soluble epoxide hydrolase complexed with N-cycloheptyl-1-(mesitylsulfonyl)piperidine-4-carboxamide. | | Descriptor: | Bifunctional epoxide hydrolase 2, MAGNESIUM ION, N-cycloheptyl-1-[(2,4,6-trimethylphenyl)sulfonyl]piperidine-4-carboxamide, ... | | Authors: | Pecic, S, Pakhomova, S, Newcomer, M.E, Morisseau, C, Hammock, B.D, Zhu, Z, Deng, S. | | Deposit date: | 2012-09-26 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Synthesis and structure-activity relationship of piperidine-derived non-urea soluble epoxide hydrolase inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3VEX
 
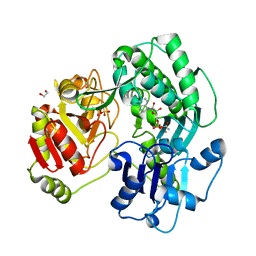 | | Crystal structure of the O-carbamoyltransferase TobZ H14N variant in complex with carbamoyl adenylate intermediate | | Descriptor: | 1,2-ETHANEDIOL, 5'-O-[(S)-(carbamoyloxy)(hydroxy)phosphoryl]adenosine, FE (II) ION, ... | | Authors: | Parthier, C, Stubbs, M.T, Goerlich, S, Jaenecke, F. | | Deposit date: | 2012-01-09 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The O-Carbamoyltransferase TobZ Catalyzes an Ancient Enzymatic Reaction.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
4N3C
 
 | |
2Z9U
 
 | | Crystal structure of pyridoxamine-pyruvate aminotransferase from Mesorhizobium loti at 2.0 A resolution | | Descriptor: | Aspartate aminotransferase, GLYCEROL, SULFATE ION | | Authors: | Yoshikane, Y, Yokochi, N, Yamasaki, M, Mizutani, K, Ohnishi, K, Mikami, B, Hayashi, H, Yagi, T. | | Deposit date: | 2007-09-26 | | Release date: | 2007-11-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of pyridoxamine-pyruvate aminotransferase from Mesorhizobium loti MAFF303099
J.Biol.Chem., 283, 2008
|
|
