1K56
 
 | | OXA 10 class D beta-lactamase at pH 6.5 | | Descriptor: | OXA10 beta-lactamase, SULFATE ION | | Authors: | Golemi, D, Maveyraud, L, Vakulenko, S, Samama, J.P, Mobashery, S. | | Deposit date: | 2001-10-10 | | Release date: | 2001-11-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Critical involvement of a carbamylated lysine in catalytic function of class D beta-lactamases.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
5AQN
 
 | | Fragment-based screening of HSP70 sheds light on the functional role of ATP-binding site residues | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BAG FAMILY MOLECULAR CHAPERONE REGULATOR 1, BENZOFURO[3,2-D]PYRIMIDIN-4(3H)-ONE, ... | | Authors: | Jones, A.M, Westwood, I.M, Osborne, J.D, Matthews, T.P, Cheeseman, M.D, Rowlands, M.G, Jeganathan, F, Burke, R, Lee, D, Kadi, N, Liu, M, Richards, M, McAndrew, C, Yahya, N, Dobson, S.E, Jones, K, Workman, P, Collins, I, van Montfort, R.L.M. | | Deposit date: | 2015-09-22 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A fragment-based approach applied to a highly flexible target: Insights and challenges towards the inhibition of HSP70 isoforms.
Sci Rep, 6, 2016
|
|
1K57
 
 | | OXA 10 class D beta-lactamase at pH 6.0 | | Descriptor: | BETA LACTAMASE OXA-10, SULFATE ION | | Authors: | Golemi, D, Maveyraud, L, Vakulenko, S, Samama, J.P, Mobashery, S. | | Deposit date: | 2001-10-10 | | Release date: | 2001-11-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Critical involvement of a carbamylated lysine in catalytic function of class D beta-lactamases.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
6ZHN
 
 | | 3D electron diffraction structure of thaumatin from Thaumatococcus daniellii | | Descriptor: | CHLORIDE ION, Thaumatin-1 | | Authors: | Blum, T, Housset, D, Clabbers, M.T.B, van Genderen, E, Schoehn, G, Ling, W.L, Abrahams, J.P. | | Deposit date: | 2020-06-23 | | Release date: | 2021-01-27 | | Last modified: | 2024-10-16 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.76 Å) | | Cite: | Statistically correcting dynamical electron scattering improves the refinement of protein nanocrystals, including charge refinement of coordinated metals.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5BMF
 
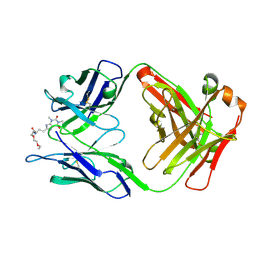 | | Crystal Structure of a Theophylline binding antibody Fab fragment | | Descriptor: | 2-(5-{1-[1-(1,3-dimethyl-2,6-dioxo-2,3,6,7-tetrahydro-1H-purin-8-yl)-4,15-dioxo-8,11-dioxa-5,14-diazaicosan-20-yl]-3,3-dimethyl-6-sulfo-1,3-dihydro-2H-indol-2-ylidene}penta-1,3-dien-1-yl)-1-ethyl-3,3-dimethyl-3H-indolium-5-sulfonate, Fab fragment heavy chain, Fab fragment light chain | | Authors: | Bujotzek, A, Fuchs, A, Changtao, Q, Klostermann, S, Benz, J, Antes, I, Dengl, S, Hoffmann, E, Georges, G. | | Deposit date: | 2015-05-22 | | Release date: | 2015-07-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | MoFvAb: Modeling the Fv region of antibodies.
Mabs, 7, 2015
|
|
7S85
 
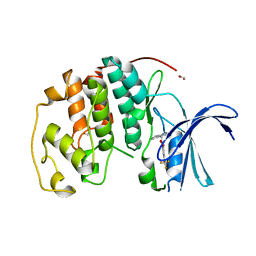 | | Crystal structure of CDK2 liganded with compound WN316 | | Descriptor: | 1,2-ETHANEDIOL, 2-{[2-(1H-indol-3-yl)ethyl]amino}-5-(trifluoromethoxy)benzoic acid, Cyclin-dependent kinase 2 | | Authors: | Sun, L, Schonbrunn, E. | | Deposit date: | 2021-09-17 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Screening through Lead Optimization of High Affinity, Allosteric Cyclin-Dependent Kinase 2 (CDK2) Inhibitors as Male Contraceptives That Reduce Sperm Counts in Mice.
J.Med.Chem., 66, 2023
|
|
1K9D
 
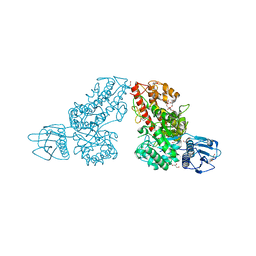 | | The 1.7 A crystal structure of alpha-D-glucuronidase, a family-67 glycoside hydrolase from Bacillus stearothermophilus T-1 | | Descriptor: | GLYCEROL, alpha-D-glucuronidase | | Authors: | Golan, G, Shallom, D, Teplitsky, A, Zaide, G, Shulami, S, Baasov, T, Stojanoff, V, Thompson, A, Shoham, Y, Shoham, G. | | Deposit date: | 2001-10-29 | | Release date: | 2002-10-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structures of Geobacillus stearothermophilus {alpha}-Glucuronidase Complexed with Its Substrate and Products: MECHANISTIC IMPLICATIONS.
J.Biol.Chem., 279, 2004
|
|
5OVL
 
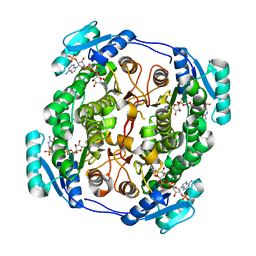 | | crystal structure of MabA bound to NADP+ from M. smegmatis | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] reductase FabG, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Kussau, T, Van Wyk, N, Viljoen, A, Olieric, V, Flipo, M, Kremer, L, Blaise, M. | | Deposit date: | 2017-08-29 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural rearrangements occurring upon cofactor binding in the Mycobacterium smegmatis beta-ketoacyl-acyl carrier protein reductase MabA.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6Z1H
 
 | | Ancestral glycosidase (family 1) | | Descriptor: | ANCESTRAL RECONSTRUCTED GLYCOSIDASE, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Gavira, J.A, Risso, V.A, Sanchez-Ruiz, J.M, Gamiz-Arco, G, Gutierrez-Rus, L, Ibarra-Molero, B, Hoshino, Y, Petrovic, D, Romero-Rivera, A, Seelig, B, Kamerlin, S.C.L, Gaucher, E.A. | | Deposit date: | 2020-05-13 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Heme-binding enables allosteric modulation in an ancient TIM-barrel glycosidase.
Nat Commun, 12, 2021
|
|
1ZFM
 
 | | AGC Duplex B-DNA | | Descriptor: | 5'-D(*CP*CP*GP*CP*TP*AP*GP*CP*GP*G)-3' | | Authors: | Hays, F.A, Teegarden, A.T, Jones, Z.J.R, Harms, M, Raup, D, Watson, J, Cavaliere, E, Ho, P.S. | | Deposit date: | 2005-04-20 | | Release date: | 2005-05-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | How sequence defines structure: a crystallographic map of DNA structure and conformation.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
5BRM
 
 | |
5B6X
 
 | | A three dimensional movie of structural changes in bacteriorhodopsin: structure obtained 760 ns after photoexcitation | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Royant, A, Nango, E, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-06-02 | | Release date: | 2016-12-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
1Z79
 
 | |
1Z7Q
 
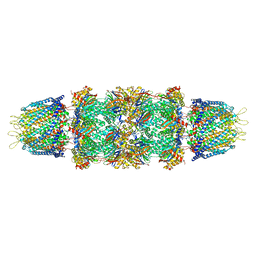 | | Crystal structure of the 20s proteasome from yeast in complex with the proteasome activator PA26 from Trypanosome brucei at 3.2 angstroms resolution | | Descriptor: | Potential proteasome component C5, Proteasome component C1, Proteasome component C11, ... | | Authors: | Forster, A, Whitby, F.G, Hill, C.P. | | Deposit date: | 2005-03-26 | | Release date: | 2005-08-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | The 1.9 A structure of a proteasome-11S activator complex and implications for proteasome-PAN/PA700 interactions.
Mol.Cell, 18, 2005
|
|
5BV0
 
 | |
7S7P
 
 | |
6ZGM
 
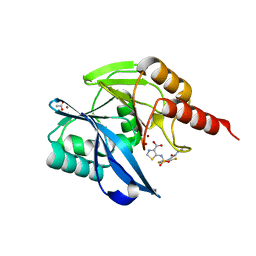 | | Crystal Structure of the VIM-2 Acquired Metallo-beta-Lactamase in Complex with the thiazolecarboxylate inhibitor ANT2681 | | Descriptor: | 5-[[4-(carbamimidamidocarbamoylamino)-3,5-bis(fluoranyl)phenyl]sulfonylamino]-1,3-thiazole-4-carboxylic acid, ACETATE ION, Metallo-beta-lactamase VIM-2-like protein, ... | | Authors: | Docquier, J.D, Pozzi, C, Marcoccia, F, De Luca, F, Benvenuti, M, Mangani, S. | | Deposit date: | 2020-06-19 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | ANT2681: SAR Studies Leading to the Identification of a Metallo-beta-lactamase Inhibitor with Potential for Clinical Use in Combination with Meropenem for the Treatment of Infections Caused by NDM-ProducingEnterobacteriaceae.
Acs Infect Dis., 6, 2020
|
|
7SI1
 
 | |
1KIO
 
 | | SOLUTION STRUCTURE OF THE SMALL SERINE PROTEASE INHIBITOR SGCI[L30R, K31M] | | Descriptor: | SERINE PROTEASE INHIBITOR I | | Authors: | Gaspari, Z, Patthy, A, Graf, L, Perczel, A. | | Deposit date: | 2001-12-03 | | Release date: | 2001-12-12 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Comparative structure analysis of proteinase inhibitors from the desert locust, Schistocerca gregaria.
Eur.J.Biochem., 269, 2002
|
|
5AQU
 
 | | Fragment-based screening of HSP70 sheds light on the functional role of ATP-binding site residues | | Descriptor: | (1S,2R,3R,5R)-3-(HYDROXYMETHYL)-5-((5-METHOXYQUINAZOLIN-4-YL)AMINO)CYCLOPENTANE-1,2-DIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BAG FAMILY MOLECULAR CHAPERONE REGULATOR 1, ... | | Authors: | Jones, A.M, Westwood, I.M, Osborne, J.D, Matthews, T.P, Cheeseman, M.D, Rowlands, M.G, Jeganathan, F, Burke, R, Lee, D, Kadi, N, Liu, M, Richards, M, McAndrew, C, Yahya, N, Dobson, S.E, Jones, K, Workman, P, Collins, I, van Montfort, R.L.M. | | Deposit date: | 2015-09-22 | | Release date: | 2016-10-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | A fragment-based approach applied to a highly flexible target: Insights and challenges towards the inhibition of HSP70 isoforms.
Sci Rep, 6, 2016
|
|
7SHV
 
 | | Crystal structure of BRAF kinase domain bound to GDC0879 | | Descriptor: | 2-{4-[(1E)-1-(hydroxyimino)-2,3-dihydro-1H-inden-5-yl]-3-(pyridin-4-yl)-1H-pyrazol-1-yl}ethanol, CHLORIDE ION, Serine/threonine-protein kinase B-raf | | Authors: | Kung, J.E, Sudhamsu, J. | | Deposit date: | 2021-10-11 | | Release date: | 2022-10-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Clearing the Path to Rapid High-Quality Protein Purification
To Be Published
|
|
5AQG
 
 | | Fragment-based screening of HSP70 sheds light on the functional role of ATP-binding site residues | | Descriptor: | (2R,3R,4S,5R)-2-(3-AMINO-5-METHYL-1,4,5,6,8-PENTAAZAACENAPHTHYLEN-1(5H)-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BAG FAMILY MOLECULAR CHAPERONE REGULATOR 1, ... | | Authors: | Jones, A.M, Westwood, I.M, Osborne, J.D, Matthews, T.P, Cheeseman, M.D, Rowlands, M.G, Jeganathan, F, Burke, R, Lee, D, Kadi, N, Liu, M, Richards, M, McAndrew, C, Yahya, N, Dobson, S.E, Jones, K, Workman, P, Collins, I, van Montfort, R.L.M. | | Deposit date: | 2015-09-22 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | A fragment-based approach applied to a highly flexible target: Insights and challenges towards the inhibition of HSP70 isoforms.
Sci Rep, 6, 2016
|
|
7SJ3
 
 | | Structure of CDK4-Cyclin D3 bound to abemaciclib | | Descriptor: | Cyclin-dependent kinase 4, G1/S-specific cyclin-D3, N-{5-[(4-ethylpiperazin-1-yl)methyl]pyridin-2-yl}-5-fluoro-4-[4-fluoro-2-methyl-1-(propan-2-yl)-1H-benzimidazol-6-yl]py rimidin-2-amine | | Authors: | Hilgers, M.T, Pelletier, L.A. | | Deposit date: | 2021-10-15 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure of CDK4-Cyclin D3 bound to abemaciclib
To Be Published
|
|
5AYP
 
 | |
1KD8
 
 | | X-RAY STRUCTURE OF THE COILED COIL GCN4 ACID BASE HETERODIMER ACID-d12Ia16V BASE-d12La16L | | Descriptor: | GCN4 ACID BASE HETERODIMER ACID-d12Ia16V, GCN4 ACID BASE HETERODIMER BASE-d12La16L | | Authors: | Keating, A.E, Malashkevich, V.N, Tidor, B, Kim, P.S. | | Deposit date: | 2001-11-12 | | Release date: | 2001-11-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Side-chain repacking calculations for predicting structures and stabilities of heterodimeric coiled coils.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
