8TE5
 
 | |
8TE4
 
 | |
8TE3
 
 | |
8TE2
 
 | |
8TE1
 
 | |
8TE0
 
 | |
8TDZ
 
 | |
8TDY
 
 | |
8TDX
 
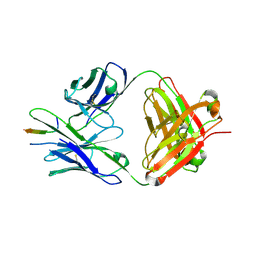 | | TRNM-b.01 in complex with HIV Env fusion peptide | | Descriptor: | Env Fusion Peptide, TRNM-b.01 Fab Heavy Chain, TRNM-b.01 Fab Light Chain | | Authors: | Olia, A.S, Kwong, P.D. | | Deposit date: | 2023-07-05 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Broad and potent HIV-1 neutralization in fusion peptide-primed SHIV-boosted macaques
To Be Published
|
|
8TDW
 
 | | ssRNA bound SAMHD1 T open | | Descriptor: | Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, FE (III) ION, RNA (5'-R(P*CP*CP*GP*AP*CP*C)-3'), ... | | Authors: | Sung, M, Huynh, K, Han, S. | | Deposit date: | 2023-07-05 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Guanine-containing ssDNA and RNA induce dimeric and tetrameric structural forms of SAMHD1.
Nucleic Acids Res., 51, 2023
|
|
8TDV
 
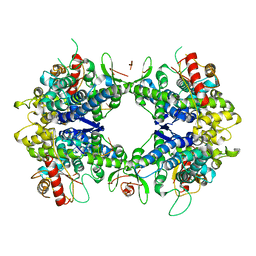 | | ssRNA bound SAMHD1 T closed | | Descriptor: | Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, FE (III) ION, RNA (5'-R(P*CP*CP*GP*AP*CP*CP*C)-3'), ... | | Authors: | Sung, M, Huynh, K, Han, S. | | Deposit date: | 2023-07-05 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Guanine-containing ssDNA and RNA induce dimeric and tetrameric structural forms of SAMHD1.
Nucleic Acids Res., 51, 2023
|
|
8TDU
 
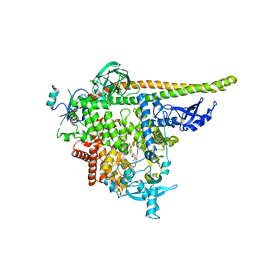 | | STX-478, a Mutant-Selective, Allosteric Inhibitor bound to PI3Kalpha | | Descriptor: | N-(2-aminopyrimidin-5-yl)-N'-[(1R)-1-(5,7-difluoro-3-methyl-1-benzofuran-2-yl)-2,2,2-trifluoroethyl]urea, N~2~-{(4S,11aP)-2-[(4S)-4-(difluoromethyl)-2-oxo-1,3-oxazolidin-3-yl]-5,6-dihydroimidazo[1,2-d][1,4]benzoxazepin-9-yl}-L-alaninamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, ... | | Authors: | Hilbert, B.J, Brooijmans, N, Buckbinder, L, St.Jean Jr, D.J. | | Deposit date: | 2023-07-05 | | Release date: | 2023-09-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | STX-478, a Mutant-Selective, Allosteric PI3K alpha Inhibitor Spares Metabolic Dysfunction and Improves Therapeutic Response in PI3K alpha-Mutant Xenografts.
Cancer Discov, 13, 2023
|
|
8TDT
 
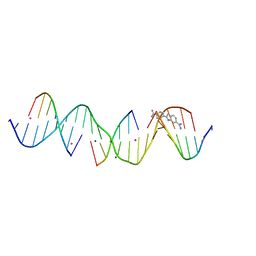 | | Sequence specific (AATT) orientation of DAPI molecules at a unique minor groove binding site (position2) within a self-assembled 3D DNA lattice (4x6) | | Descriptor: | 6-AMIDINE-2-(4-AMIDINO-PHENYL)INDOLE, CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*TP*GP*AP*CP*GP*GP*AP*AP*AP*TP*TP*A)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2023-07-04 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.007 Å) | | Cite: | Site-Specific Arrangement and Structure Determination of Minor Groove Binding Molecules in Self-Assembled Three-Dimensional DNA Crystals.
J.Am.Chem.Soc., 145, 2023
|
|
8TDR
 
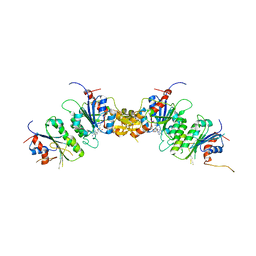 | |
8TDQ
 
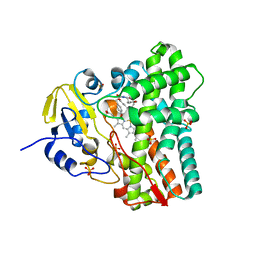 | | SFX-XFEL structure of CYP121 cocrystallized with substrate cYY | | Descriptor: | (3S,6S)-3,6-bis(4-hydroxybenzyl)piperazine-2,5-dione, Mycocyclosin synthase, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Nguyen, R.C, Dasgupta, M, Bhowmick, A, Kern, J.F, Liu, A. | | Deposit date: | 2023-07-04 | | Release date: | 2023-11-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | In Situ Structural Observation of a Substrate- and Peroxide-Bound High-Spin Ferric-Hydroperoxo Intermediate in the P450 Enzyme CYP121.
J.Am.Chem.Soc., 145, 2023
|
|
8TDP
 
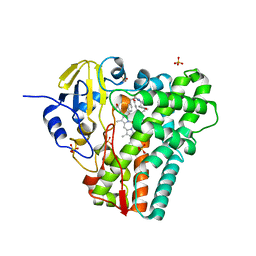 | | Time-resolved SFX-XFEL crystal structure of CYP121 bound with cYY reacted with peracetic acid for 200 milliseconds | | Descriptor: | (3S,6S)-3,6-bis(4-hydroxybenzyl)piperazine-2,5-dione, HYDROGEN PEROXIDE, Mycocyclosin synthase, ... | | Authors: | Nguyen, R.C, Dasgupta, M, Bhowmick, A, Kern, J.F, Liu, A. | | Deposit date: | 2023-07-04 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | In Situ Structural Observation of a Substrate- and Peroxide-Bound High-Spin Ferric-Hydroperoxo Intermediate in the P450 Enzyme CYP121.
J.Am.Chem.Soc., 145, 2023
|
|
8TDO
 
 | |
8TDN
 
 | |
8TDM
 
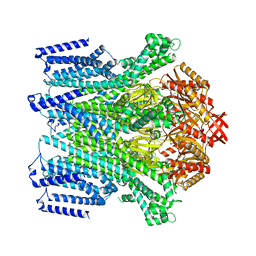 | | Cryo-EM structure of AtMSL10-K539E | | Descriptor: | Mechanosensitive ion channel protein 10 | | Authors: | Zhang, J, Yuan, P. | | Deposit date: | 2023-07-03 | | Release date: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Open structure and gating of the Arabidopsis mechanosensitive ion channel MSL10.
Nat Commun, 14, 2023
|
|
8TDL
 
 | |
8TDK
 
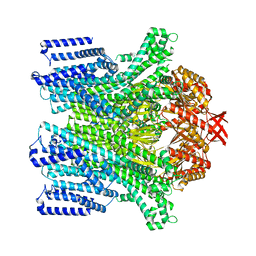 | | Cryo-EM structure of AtMSL10-G556V | | Descriptor: | Mechanosensitive ion channel protein 10 | | Authors: | Zhang, J, Yuan, P. | | Deposit date: | 2023-07-03 | | Release date: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Open structure and gating of the Arabidopsis mechanosensitive ion channel MSL10.
Nat Commun, 14, 2023
|
|
8TDJ
 
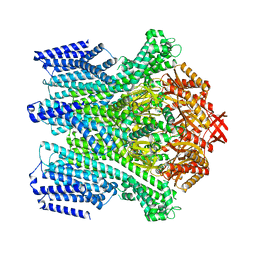 | |
8TDI
 
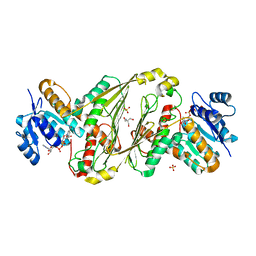 | | Structure of P2B11 Glucuronide-3-dehydrogenase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, P2B11 Glucuronide-3-dehydrogenase, ... | | Authors: | Lazarski, A.C, Worrall, L.J, Strynadka, N.C.J. | | Deposit date: | 2023-07-03 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An alternative broad-specificity pathway for glycan breakdown in bacteria.
Nature, 631, 2024
|
|
8TDH
 
 | |
8TDF
 
 | |
