3A4Y
 
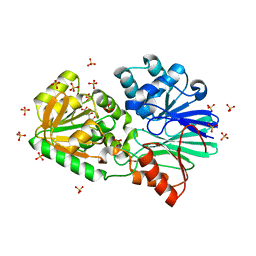 | | Crystal Structure of H61A mutant TTHA0252 from Thermus thermophilus HB8 | | Descriptor: | CITRATE ANION, Ribonuclease TTHA0252, SULFATE ION, ... | | Authors: | Ishikawa, H, Nakagawa, N, Kuramitsu, S, Yokoyama, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-07-22 | | Release date: | 2009-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of H61A mutant TTHA0252 from Thermus thermophilus HB8
to be published
|
|
6EDU
 
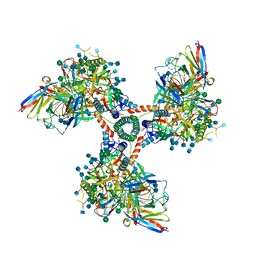 | |
6EAM
 
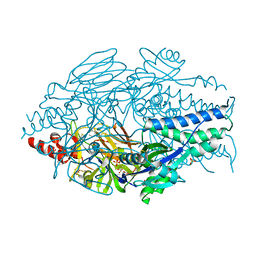 | |
6ERG
 
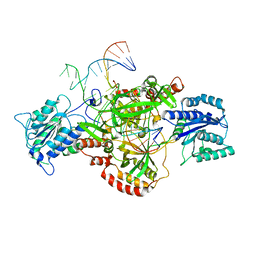 | | Complex of XLF and heterodimer Ku bound to DNA | | Descriptor: | DNA (21-MER), DNA (34-MER), Non-homologous end-joining factor 1, ... | | Authors: | Nemoz, C, Legrand, P, Ropars, V, Charbonnier, J.B. | | Deposit date: | 2017-10-18 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | XLF and APLF bind Ku80 at two remote sites to ensure DNA repair by non-homologous end joining.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6EOU
 
 | |
8QYQ
 
 | | Beta-cardiac myosin S1 fragment in the pre-powerstroke state complexed to Mavacamten | | Descriptor: | 6-[[(1~{S})-1-phenylethyl]amino]-3-propan-2-yl-1~{H}-pyrimidine-2,4-dione, ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Robert-Paganin, J, Kikuti, C, Auguin, D, Rety, S, David, A, Houdusse, A. | | Deposit date: | 2023-10-26 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Omecamtiv mecarbil and Mavacamten target the same myosin pocket despite antagonistic effects in heart contraction.
Biorxiv, 2023
|
|
8QBJ
 
 | | Structure of mBaoJin at pH 4.6 | | Descriptor: | CHLORIDE ION, mBaoJin | | Authors: | Samygina, V.R, Vlaskina, A.V, Gabdulkhakov, A, Subach, O.M, Subach, F.V. | | Deposit date: | 2023-08-24 | | Release date: | 2023-12-27 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bright and stable monomeric green fluorescent protein derived from StayGold.
Nat.Methods, 21, 2024
|
|
8Q5B
 
 | |
6EAD
 
 | |
6EAF
 
 | |
6EHX
 
 | |
8Q79
 
 | | Structure of mBaoJin at pH 6.5 | | Descriptor: | CHLORIDE ION, mBaoJin | | Authors: | Samygina, V.R, Subach, O.M, Vlaskina, A.V, Nikolaeva, A.Y, Borshchevsky, V, Qin, W, Subach, F.V. | | Deposit date: | 2023-08-15 | | Release date: | 2023-12-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Bright and stable monomeric green fluorescent protein derived from StayGold.
Nat.Methods, 21, 2024
|
|
6ELO
 
 | |
6EMG
 
 | |
6E7H
 
 | |
6EOS
 
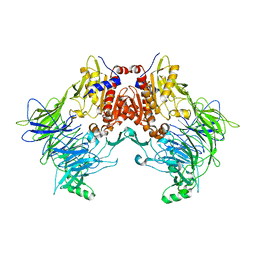 | | DPP8 - Apo, space group 19 | | Descriptor: | Dipeptidyl peptidase 8 | | Authors: | Ross, B.R, Huber, R. | | Deposit date: | 2017-10-10 | | Release date: | 2018-02-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures and mechanism of dipeptidyl peptidases 8 and 9, important players in cellular homeostasis and cancer.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8Q5Q
 
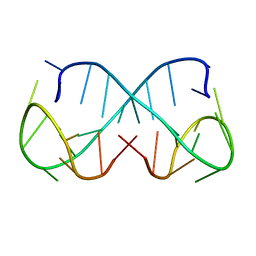 | | d(ATTTC)3 dimeric structure | | Descriptor: | DNA (5'-D(*AP*TP*TP*TP*(DNR)P*AP*TP*TP*TP*CP*AP*TP*TP*TP*C)-3') | | Authors: | Trajkovski, M, Pastore, A, Plavec, J. | | Deposit date: | 2023-08-09 | | Release date: | 2024-02-07 | | Last modified: | 2024-03-06 | | Method: | SOLUTION NMR | | Cite: | Dimeric structures of DNA ATTTC repeats promoted by divalent cations.
Nucleic Acids Res., 52, 2024
|
|
6EAK
 
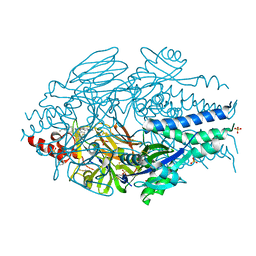 | |
6EDH
 
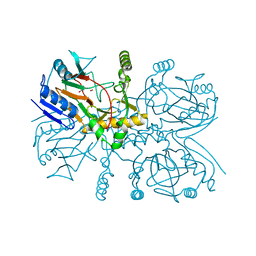 | | Taurine:2OG dioxygenase (TauD) bound to the vanadyl ion, taurine, and succinate | | Descriptor: | 2-AMINOETHANESULFONIC ACID, ACETATE ION, Alpha-ketoglutarate-dependent taurine dioxygenase, ... | | Authors: | Davis, K.M, Altmyer, M, Boal, A.K. | | Deposit date: | 2018-08-09 | | Release date: | 2019-08-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.73000407 Å) | | Cite: | Structure of a Ferryl Mimic in the Archetypal Iron(II)- and 2-(Oxo)-glutarate-Dependent Dioxygenase, TauD.
Biochemistry, 58, 2019
|
|
8QNV
 
 | |
8QNT
 
 | |
6ELN
 
 | | Estimation of relative drug-target residence times by random acceleration molecular dynamics simulation | | Descriptor: | 4-[4-(4-methoxyphenyl)-5-methyl-1H-pyrazol-3-yl]benzene-1,3-diol, Heat shock protein HSP 90-alpha, SULFATE ION | | Authors: | Musil, D, Lehmann, M, Eggenweiler, H.-M. | | Deposit date: | 2017-09-29 | | Release date: | 2018-05-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Estimation of Drug-Target Residence Times by tau-Random Acceleration Molecular Dynamics Simulations.
J Chem Theory Comput, 14, 2018
|
|
6EMN
 
 | | HtxB from Pseudomonas stutzeri in complex with phosphite to 1.25 A resolution | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Probable phosphite transport system-binding protein HtxB, ... | | Authors: | Bisson, C, Robertson, A.J, Hitchcock, A, Adams, N.B. | | Deposit date: | 2017-10-03 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Phosphite binding by the HtxB periplasmic binding protein depends on the protonation state of the ligand.
Sci Rep, 9, 2019
|
|
6EO9
 
 | | Crystal structure of thrombin in complex with a novel glucose-conjugated potent inhibitor | | Descriptor: | DIMETHYL SULFOXIDE, Hirudin variant-2, N-(2-{[5-(5-chlorothiophen-2-yl)-1,2-oxazol-3-yl]methoxy}-6-{3-[(2,3,4,6-tetra-O-acetyl-beta-D-glucopyranosyl)oxy]propoxy}phenyl)-1-(propan-2-yl)piperidine-4-carboxamide, ... | | Authors: | Belviso, B.D, Caliandro, R, Aresta, B.M, De Candia, M, Altomare, C.D. | | Deposit date: | 2017-10-09 | | Release date: | 2017-12-13 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | How a beta-D-glucoside side chain enhances binding affinity to thrombin of inhibitors bearing 2-chlorothiophene as P1 moiety: crystallography, fragment deconstruction study, and evaluation of antithrombotic properties.
J. Med. Chem., 57, 2014
|
|
6EAE
 
 | |
