6OBS
 
 | | PP1 Y134K | | Descriptor: | GLYCEROL, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Choy, M.S, Moon, T.M, Bray, J.A, Archuleta, T.L, Shi, W, Peti, W, Page, R. | | Deposit date: | 2019-03-21 | | Release date: | 2019-09-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | SDS22 selectively recognizes and traps metal-deficient inactive PP1.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
8E4R
 
 | |
8UPI
 
 | | Structure of a periplasmic peptide binding protein from Mesorhizobium sp. AP09 bound to aminoserine | | Descriptor: | 1,2-ETHANEDIOL, AMINOSERINE, CALCIUM ION, ... | | Authors: | Frkic, R.L, Smith, O.B, Rahman, M, Kaczmarski, J.A, Jackson, C.J. | | Deposit date: | 2023-10-22 | | Release date: | 2023-11-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Identification and Characterization of a Bacterial Periplasmic Solute Binding Protein That Binds l-Amino Acid Amides.
Biochemistry, 63, 2024
|
|
6FES
 
 | | Crystal structure of novel repeat protein BRIC2 fused to DARPin D12 | | Descriptor: | 1,2-ETHANEDIOL, D12_BRIC2, a synthetic protein,D12_BRIC2, ... | | Authors: | ElGamacy, M, Coles, M, Ernst, P, Zhu, H, Hartmann, M.D, Plueckthun, A, Lupas, A. | | Deposit date: | 2018-01-03 | | Release date: | 2018-09-05 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | An Interface-Driven Design Strategy Yields a Novel, Corrugated Protein Architecture.
ACS Synth Biol, 7, 2018
|
|
9AV1
 
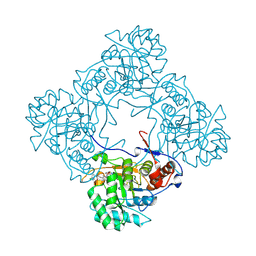 | | Crystal structure of E. coli GuaB dCBS with inhibitor GNE9123 | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase, N-(6-chloropyridin-3-yl)-N~2~-(1,4-dihydro-2H-pyrano[3,4-c]quinolin-9-yl)-L-alaninamide | | Authors: | Harris, S.F, Wu, P. | | Deposit date: | 2024-03-01 | | Release date: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of GuaB inhibitors with efficacy against Acinetobacter baumannii infection.
Mbio, 15, 2024
|
|
7POY
 
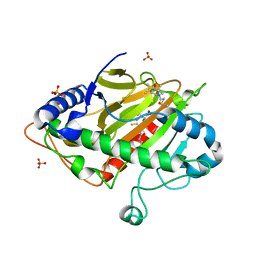 | | Spin labeled IPNS S55C variant in complex with Fe, ACV and NO | | Descriptor: | FE (III) ION, Isopenicillin N synthase, L-D-(A-AMINOADIPOYL)-L-CYSTEINYL-D-VALINE, ... | | Authors: | Rabe, P, Clifton, I, Walla, C, Schofield, C.J. | | Deposit date: | 2021-09-10 | | Release date: | 2022-07-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Spectroscopic studies reveal details of substrate-induced conformational changes distant from the active site in isopenicillin N synthase.
J.Biol.Chem., 298, 2022
|
|
6QW7
 
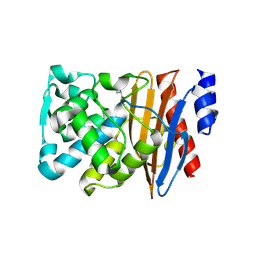 | | Crystal structure of L2 complexed with relebactam (16 hour soak) | | Descriptor: | (2S,5R)-1-formyl-N-(piperidin-4-yl)-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase, D-SERINE | | Authors: | Tooke, C.L, Hinchliffe, P, Spencer, J. | | Deposit date: | 2019-03-05 | | Release date: | 2019-08-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Molecular Basis of Class A beta-Lactamase Inhibition by Relebactam.
Antimicrob.Agents Chemother., 63, 2019
|
|
6QVP
 
 | |
6XSZ
 
 | |
9JYS
 
 | | Crystal structure of the PIN1 and fragment 5 complex. | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, 4-(aminomethyl)-N,N-dimethyl-aniline, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, ... | | Authors: | Xiao, Q.J, Wu, T.T, Shu, H.L, Qin, W.M. | | Deposit date: | 2024-10-12 | | Release date: | 2025-10-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Fragment-Based Compound Screening Based on PIN1 Crystals
To Be Published
|
|
8V98
 
 | | GII.24 Loreto 1972 norovirus protruding domain | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Capsid protein VP1 | | Authors: | Kher, G, Prewitt, A, Pancera, M, Hansman, G. | | Deposit date: | 2023-12-07 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Development of a broad-spectrum therapeutic Fc-nanobody for human noroviruses.
J.Virol., 98, 2024
|
|
9AVD
 
 | |
367D
 
 | | 1.2 A STRUCTURE DETERMINATION OF THE D(CG(5-BRU)ACG)2/5-BROMO-9-AMINO-DACA COMPLEX | | Descriptor: | 5'-D(*CP*GP*(BRU)P*AP*CP*G)-3', 5-BROMO-9-AMINO-N-ETHYL(DIAMINOMETHYL)ACRIDINE-4-CARBOXAMIDE, BROMIDE ION | | Authors: | Todd, A.K, Adams, A, Thorpe, J.H, Denny, W.A, Cardin, C.J. | | Deposit date: | 1997-12-19 | | Release date: | 2003-03-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Major groove binding and 'DNA-induced' fit in the intercalation of a derivative of the mixed topoisomerase I/II poison N-(2-(dimethlyamino)ethyl)acridine-4-carboxamide (DACA) into DNA: X-ray structure complexed to d(CG(5Br-U)ACG)2 at 1.3-angstrom resolution
J.Med.Chem., 42, 1999
|
|
5L9I
 
 | |
6N53
 
 | | Crystal structure of human uridine-cytidine kinase 2 complexed with 2'-azidouridine monophosphate | | Descriptor: | 2'-deoxy-2'-triaza-1,2-dien-2-ium-1-yl-uridine-5'-monophosphate, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Cuthbert, B.J, Nainar, S, Spitale, R.C, Goulding, C.W. | | Deposit date: | 2018-11-21 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An optimized chemical-genetic method for cell-specific metabolic labeling of RNA.
Nat.Methods, 17, 2020
|
|
9S87
 
 | |
9S8S
 
 | |
9S80
 
 | |
6PKU
 
 | | Guinea pig N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase (NAGPA) catalytic domain (C51S C221S) in complex with N-acetyl-alpha-D-glucosamine (alpha-GlcNAc) and mannose 6-phosphate (M6P) | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-O-phosphono-alpha-D-mannopyranose, ... | | Authors: | Gorelik, A, Illes, K, Nagar, B. | | Deposit date: | 2019-06-29 | | Release date: | 2020-02-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Crystal Structure of the Mannose-6-Phosphate Uncovering Enzyme.
Structure, 28, 2020
|
|
7OR2
 
 | | Crystal structure of UDP-N-acetylenolpyruvoylglucosamine reductase (MurB) from Pseudomonas aeruginosa in complex with FAD and a pyrazole derivative (fragment 4) | | Descriptor: | 5-methyl-1-phenyl-pyrazole-4-carboxylic acid, FLAVIN-ADENINE DINUCLEOTIDE, UDP-N-acetylenolpyruvoylglucosamine reductase | | Authors: | Acebron-Garcia de Eulate, M, Blundell, T.L, Kim, S.Y, Mendes, V, Abell, C. | | Deposit date: | 2021-06-04 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of Novel Inhibitors of Uridine Diphosphate- N -Acetylenolpyruvylglucosamine Reductase (MurB) from Pseudomonas aeruginosa , an Opportunistic Infectious Agent Causing Death in Cystic Fibrosis Patients.
J.Med.Chem., 65, 2022
|
|
6MUH
 
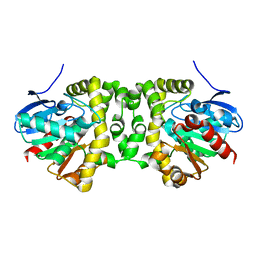 | | Fluoroacetate dehalogenase, room temperature structure solved by serial 1 degree oscillation crystallography | | Descriptor: | Fluoroacetate dehalogenase | | Authors: | Finke, A.D, Wierman, J.L, Pare-Labrosse, O, Sarrachini, A, Besaw, J, Mehrabi, P, Gruner, S.M, Miller, R.J.D. | | Deposit date: | 2018-10-23 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fixed-target serial oscillation crystallography at room temperature.
IUCrJ, 6, 2019
|
|
8QFM
 
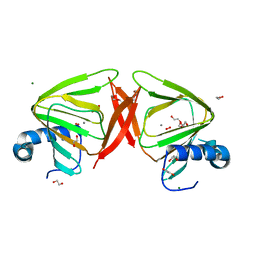 | |
8Y59
 
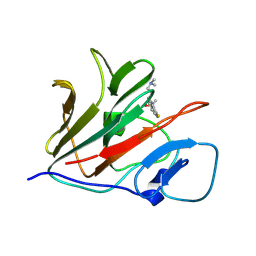 | | Crystal structure of TRIM21 PRYSPRY (D355A) in complex with (S)-hydroxyl-acepromazine. | | Descriptor: | (1~{S})-1-[10-[3-(dimethylamino)propyl]phenothiazin-2-yl]ethanol, E3 ubiquitin-protein ligase TRIM21 | | Authors: | Lu, P, Cheng, Y, Xue, L, Ren, X, Huang, N, Han, T. | | Deposit date: | 2024-01-31 | | Release date: | 2024-10-09 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Selective degradation of multimeric proteins by TRIM21-based molecular glue and PROTAC degraders.
Cell, 187, 2024
|
|
8Y5B
 
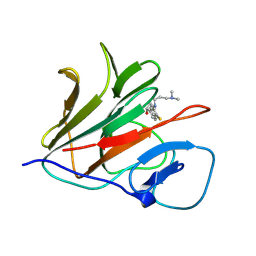 | | Crystal structure of TRIM21 PRYSPRY (D355A) in complex with (R)-hydroxyl-acepromazine. | | Descriptor: | (1~{R})-1-[10-[3-(dimethylamino)propyl]phenothiazin-2-yl]ethanol, E3 ubiquitin-protein ligase TRIM21 | | Authors: | Lu, P, Cheng, Y, Xue, L, Ren, X, Huang, N, Han, T. | | Deposit date: | 2024-01-31 | | Release date: | 2024-10-09 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Selective degradation of multimeric proteins by TRIM21-based molecular glue and PROTAC degraders.
Cell, 187, 2024
|
|
5WHM
 
 | | Crystal Structure of IclR Family Transcriptional Regulator from Brucella abortus | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CALCIUM ION, ... | | Authors: | Kim, Y, Wu, R, Tesar, C, Endres, M, Babnigg, G, Crosson, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-07-17 | | Release date: | 2017-08-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular control of gene expression byBrucellaBaaR, an IclR-type transcriptional repressor.
J. Biol. Chem., 293, 2018
|
|
