3NTA
 
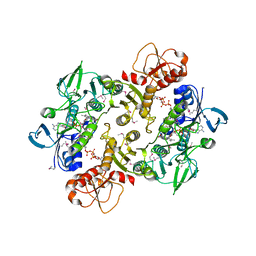 | | Structure of the Shewanella loihica PV-4 NADH-dependent persulfide reductase | | Descriptor: | CHLORIDE ION, COENZYME A, FAD-dependent pyridine nucleotide-disulphide oxidoreductase, ... | | Authors: | Sazinsky, M.H, Crane, E.J, Warner, M.D, Lukose, V, Lee, K.H. | | Deposit date: | 2010-07-03 | | Release date: | 2010-12-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Characterization of an NADH-Dependent Persulfide Reductase from Shewanella loihica PV-4: Implications for the Mechanism of Sulfur Respiration via FAD-Dependent Enzymes .
Biochemistry, 50, 2010
|
|
6R2C
 
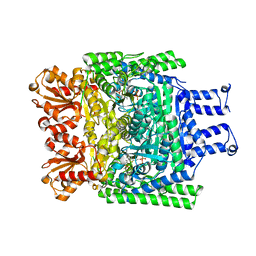 | | Crystal structure of the SucA domain of Mycobacterium smegmatis KGD after soaking with succinylphosphonate phosphonoethyl ester (PESP) | | Descriptor: | 4-[ethoxy(oxidanyl)phosphoryl]-4-oxidanylidene-butanoic acid, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Wagner, T, Alzari, P.M, Bellinzoni, M. | | Deposit date: | 2019-03-15 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Conformational transitions in the active site of mycobacterial 2-oxoglutarate dehydrogenase upon binding phosphonate analogues of 2-oxoglutarate: From a Michaelis-like complex to ThDP adducts.
J.Struct.Biol., 208, 2019
|
|
8C0J
 
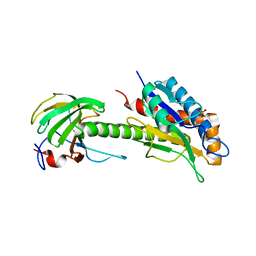 | | Structure of AmiB enzymatic domain bound to the EnvC LytM domain | | Descriptor: | Murein hydrolase activator EnvC, N-acetylmuramoyl-L-alanine amidase, PHOSPHATE ION, ... | | Authors: | Crow, A. | | Deposit date: | 2022-12-17 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (3.381 Å) | | Cite: | Activator-induced conformational changes regulate division-associated peptidoglycan amidases.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6R2B
 
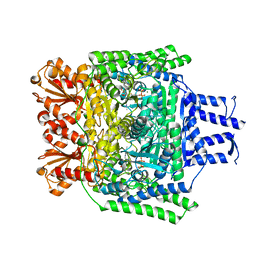 | | Crystal structure of the SucA domain of Mycobacterium smegmatis KGD after soaking with succinylphosphonate | | Descriptor: | (4~{S})-4-[(2~{R})-3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-4-methyl-5-[2-[oxidanyl(phosphonooxy)phosphoryl]oxyethyl]-2~{H}-1,3-thiazol-2-yl]-4-oxidanyl-4-phosphono-butanoic acid, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Wagner, T, Alzari, P.M, Bellinzoni, M. | | Deposit date: | 2019-03-15 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Conformational transitions in the active site of mycobacterial 2-oxoglutarate dehydrogenase upon binding phosphonate analogues of 2-oxoglutarate: From a Michaelis-like complex to ThDP adducts.
J.Struct.Biol., 208, 2019
|
|
6KU1
 
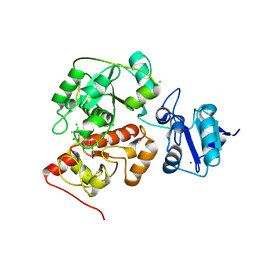 | | The structure of EanB/Y353A complex with ergothioneine | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Wu, L, Liu, P.H, Zhou, J.H. | | Deposit date: | 2019-08-30 | | Release date: | 2020-08-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Single-Step Replacement of an Unreactive C-H Bond by a C-S Bond Using Polysulfide as the Direct Sulfur Source in the Anaerobic Ergothioneine Biosynthesis
Acs Catalysis, 10, 2020
|
|
8C4P
 
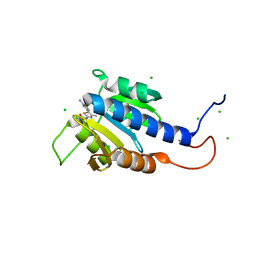 | | CdaA-compound 7 complex | | Descriptor: | CHLORIDE ION, Diadenylate cyclase, ~{N}-[5-(2-azanyl-1,3-thiazol-4-yl)-4-methyl-1,3-thiazol-2-yl]ethanamide | | Authors: | Neumann, P, Ficner, R. | | Deposit date: | 2023-01-04 | | Release date: | 2023-06-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Computer-aided design of a cyclic di-AMP synthesizing enzyme CdaA inhibitor.
Microlife, 4, 2023
|
|
3NO7
 
 | |
6KV9
 
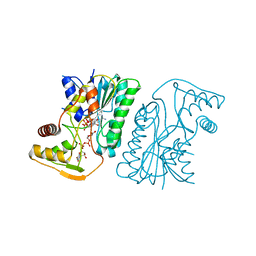 | | MoeE5 in complex with UDP-glucuronic acid and NAD | | Descriptor: | MoeE5, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, URIDINE-5'-DIPHOSPHATE-GLUCURONIC ACID | | Authors: | Ko, T.-P, Liu, W, Sun, H, Liu, W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2019-09-03 | | Release date: | 2019-11-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structure of an antibiotic-synthesizing UDP-glucuronate 4-epimerase MoeE5 in complex with substrate.
Biochem.Biophys.Res.Commun., 521, 2020
|
|
3NXF
 
 | | Robust computational design, optimization, and structural characterization of retroaldol enzymes | | Descriptor: | Retro-Aldolase, SULFATE ION | | Authors: | Althoff, E.A, Jiang, L, Wang, L, Lassila, J.K, Moody, J, Bolduc, J, Wang, Z.Z, Smith, M, Hari, S, Herschlag, D, Stoddard, B.L, Baker, D. | | Deposit date: | 2010-07-13 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural analyses of covalent enzyme-substrate analog complexes reveal strengths and limitations of de novo enzyme design.
J.Mol.Biol., 415, 2012
|
|
6R1G
 
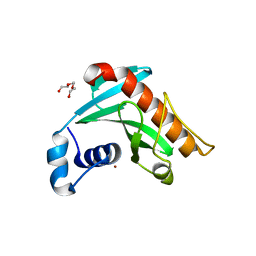 | | Crystal structure of Borrelia burgdorferi periplasmic protein BB0365 (IPLA7, p22) | | Descriptor: | Outer surface 22 kDa lipoprotein, TETRAETHYLENE GLYCOL, TRIETHYLENE GLYCOL, ... | | Authors: | Brangulis, K, Akopjana, I, Petrovskis, I, Kazaks, A, Tars, K. | | Deposit date: | 2019-03-14 | | Release date: | 2019-09-18 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural analysis of Borrelia burgdorferi periplasmic lipoprotein BB0365 involved in Lyme disease infection.
Febs Lett., 594, 2020
|
|
2XKQ
 
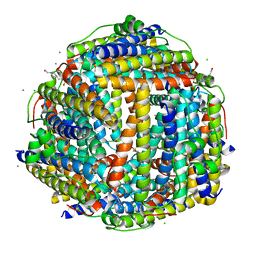 | | Crystal structure of Streptococcus suis Dpr with manganese | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Haikarainen, T, Thanassoulas, A, Stavros, P, Nounesis, G, Haataja, S, Papageorgiou, A.C. | | Deposit date: | 2010-07-12 | | Release date: | 2010-11-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Thermodynamic Characterization of Metal Ion Binding in Streptococcus Suis Dpr.
J.Mol.Biol., 405, 2011
|
|
6KYV
 
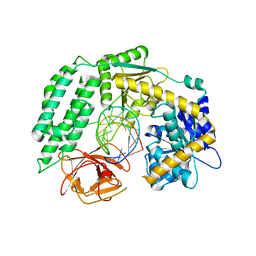 | | Crystal Structure of RIG-I and hairpin RNA with G-U wobble base pairs | | Descriptor: | Probable ATP-dependent RNA helicase DDX58, RNA (5'-R(*GP*GP*UP*AP*GP*AP*CP*GP*CP*UP*UP*CP*GP*GP*CP*GP*UP*UP*UP*GP*CP*C)-3'), ZINC ION | | Authors: | Kim, K.-H, Hwang, J, Kim, J.H, Son, K.-P, Jang, Y, Kim, M, Kang, S.-J, Lee, J.-O, Choi, B.-S. | | Deposit date: | 2019-09-20 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and biophysical properties of RIG-I bound to dsRNA with G-U wobble base pairs.
Rna Biol., 17, 2020
|
|
2YBB
 
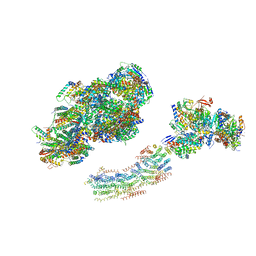 | | Fitted model for bovine mitochondrial supercomplex I1III2IV1 by single particle cryo-EM (EMD-1876) | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CALCIUM ION, CARDIOLIPIN, ... | | Authors: | Althoff, T, Mills, D.J, Popot, J.-L, Kuehlbrandt, W. | | Deposit date: | 2011-03-02 | | Release date: | 2011-10-19 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (19 Å) | | Cite: | Arrangement of Electron Transport Chain Components in Bovine Mitochondrial Supercomplex I(1)III(2)Iv(1).
Embo J., 30, 2011
|
|
2YDX
 
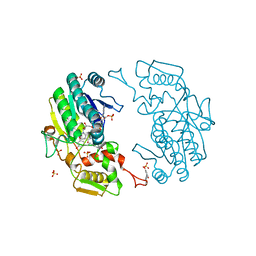 | | Crystal structure of human S-adenosylmethionine synthetase 2, beta subunit | | Descriptor: | 1,4,5,6-TETRAHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE PHOSPHATE, CALCIUM ION, METHIONINE ADENOSYLTRANSFERASE 2 SUBUNIT BETA, ... | | Authors: | Muniz, J.R.C, Shafqat, N, Pike, A.C.W, Yue, W.W, Vollmar, M, Papagriogriou, V, Roos, A, Gileadi, O, von Delft, F, Kavanagh, K.L, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Oppermann, U. | | Deposit date: | 2011-03-25 | | Release date: | 2011-04-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Insight Into S-Adenosylmethionine Biosynthesis from the Crystal Structures of the Human Methionine Adenosyltransferase Catalytic and Regulatory Subunits.
Biochem.J., 452, 2013
|
|
2YLL
 
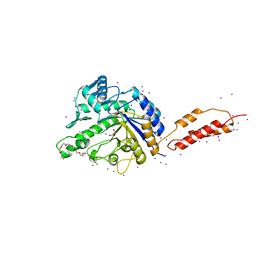 | | Inhibition of the pneumococcal virulence factor StrH and molecular insights into N-glycan recognition and hydrolysis | | Descriptor: | 1,2-ETHANEDIOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, BETA-N-ACETYLHEXOSAMINIDASE, ... | | Authors: | Pluvinage, B, Higgins, M.A, Abbott, D.W, Robb, C, Dalia, A.B, Deng, L, Weiser, J.N, Parsons, T.B, Fairbanks, A.J, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2011-06-02 | | Release date: | 2011-09-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Inhibition of the Pneumococcal Virulence Factor Strh and Molecular Insights Into N-Glycan Recognition and Hydrolysis.
Structure, 19, 2011
|
|
6R6M
 
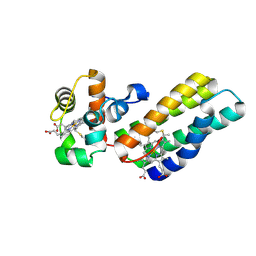 | |
3O3L
 
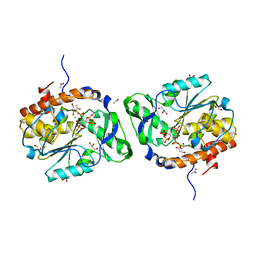 | | Structure of the PTP-like phytase from Selenomonas ruminantium in complex with myo-inositol (1,3,4,5)tetrakisphosphate | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Gruninger, R.J, Selinger, L.B, Mosimann, S.C. | | Deposit date: | 2010-07-25 | | Release date: | 2011-12-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural analysis of substrate binding in PTPLPs
To be Published
|
|
6L6L
 
 | | Structural basis of NR4A2 homodimers binding to selective Nur-responsive elements | | Descriptor: | DNA (5'-D(*AP*AP*AP*GP*GP*TP*CP*AP*AP*AP*CP*TP*GP*TP*GP*AP*CP*CP*TP*AP*T)-3'), DNA (5'-D(P*TP*AP*TP*AP*GP*GP*TP*CP*AP*CP*AP*GP*TP*TP*TP*GP*AP*CP*CP*TP*T)-3'), Nuclear receptor related 1, ... | | Authors: | Jiang, L, Chen, Y. | | Deposit date: | 2019-10-29 | | Release date: | 2019-11-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.781 Å) | | Cite: | Structural basis of binding of homodimers of the nuclear receptor NR4A2 to selective Nur-responsive DNA elements.
J.Biol.Chem., 294, 2019
|
|
6L4O
 
 | | Crystal structure of API5-FGF2 complex | | Descriptor: | Apoptosis inhibitor 5, Fibroblast growth factor 2 | | Authors: | Lee, B.I, Bong, S.M. | | Deposit date: | 2019-10-18 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Regulation of mRNA export through API5 and nuclear FGF2 interaction.
Nucleic Acids Res., 48, 2020
|
|
2YP1
 
 | | Crystallization of a 45 kDa peroxygenase- peroxidase from the mushroom Agrocybe aegerita and structure determination by SAD utilizing only the haem iron | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Piontek, K, Strittmatter, E, Ullrich, R, Plattner, D.A, Hofrichter, M. | | Deposit date: | 2012-10-29 | | Release date: | 2013-10-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural Basis of Substrate Conversion in a New Aromatic Peroxygenase: P450 Functionality with Benefits
J.Biol.Chem., 288, 2013
|
|
2YXL
 
 | | Crystal Structure of PH0851 | | Descriptor: | 450aa long hypothetical fmu protein, SINEFUNGIN | | Authors: | Hikida, Y, Kuratani, M, Bessho, Y, Ishii, R, Sekine, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-26 | | Release date: | 2008-04-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of an archaeal homologue of the bacterial Fmu/RsmB/RrmB rRNA cytosine 5-methyltransferase
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3O12
 
 | | The crystal structure of a functionally unknown protein from Saccharomyces cerevisiae. | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Uncharacterized protein YJL217W | | Authors: | Zhang, R, Tan, K, Xu, X, Cui, H, Chin, S, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-07-20 | | Release date: | 2010-09-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of a functionally unknown protein from Saccharomyces cerevisiae.
TO BE PUBLISHED
|
|
3NRS
 
 | | Crystal structure of ligand-free bifunctional folylpolyglutamate synthase/dihydrofolate synthase from yersinia pestis c092 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Dihydrofolate:folylpolyglutamate synthetase, GLYCEROL, ... | | Authors: | Nocek, B, Maltseva, N, Makowska-grzyska, M, Papazisi, L, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-06-30 | | Release date: | 2010-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of ligand-free bifunctional folylpolyglutamate synthase/dihydrofolate synthase from yersinia pestis c092
TO BE PUBLISHED
|
|
3O4Q
 
 | |
3NWY
 
 | |
