2F38
 
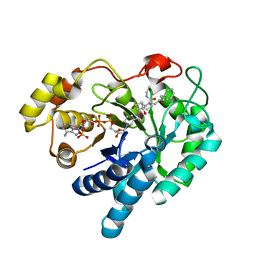 | | Crystal structure of prostaglandin F synathase containing bimatoprost | | Descriptor: | (5Z)-7-{(1R,2R,3R,5S)-3,5-DIHYDROXY-2-[(1E,3S)-3-HYDROXY-5-PHENYLPENT-1-ENYL]CYCLOPENTYL}-N-ETHYLHEPT-5-ENAMIDE, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Komoto, J, Yamada, T, Watanabe, K, Woodward, D.F, Takusagawa, F. | | Deposit date: | 2005-11-18 | | Release date: | 2006-10-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Prostaglandin F2alpha formation from prostaglandin H2 by prostaglandin F synthase (PGFS): crystal structure of PGFS containing bimatoprost.
Biochemistry, 45, 2006
|
|
8XB1
 
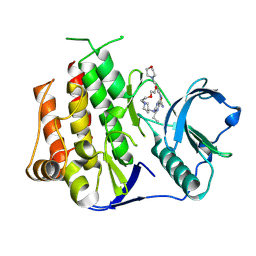 | | Crystal structure of FLT3 in complex with a Pyrazinamide Macrocycle derivative | | Descriptor: | 3^3-ethyl-5^4-morpholino-6,9-dioxa-2,4-diaza-3(2,6)-pyrazina-1(4,1)-piperidina-5(1,3)-benzenacycloundecaphane-3^5-carboxamide, Receptor-type tyrosine-protein kinase FLT3 | | Authors: | Guo, M, Chen, Y. | | Deposit date: | 2023-12-05 | | Release date: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure-Based Optimization of Pyrazinamide-Containing Macrocyclic Derivatives as Fms-like Tyrosine Kinase 3 (FLT3) Inhibitors to Overcome Clinical Mutations.
Acs Pharmacol Transl Sci, 7, 2024
|
|
9IHX
 
 | |
7VJB
 
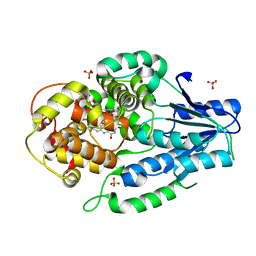 | | class II photolyase MmCPDII semiquinone to fully reduced TR-SFX studies (30 ns time-point) | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, DNA photolyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maestre-Reyna, M, Yang, C.-H, Huang, W.-C, Nango, E, Ngura Putu, E.P.G, Franz-Badur, S, Wu, W.-J, Wu, H.-Y, Wang, P.-H, Hosokawa, Y, Saft, M, Emmerich, H.-J, Liao, J.-H, Lee, C.-C, Huang, K.-F, Chang, Y.-K, Weng, J.-H, Royant, A, Gad, W, Pang, A.H, Chang, C.-W, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Yamamoto, J, Iwata, S, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2021-09-28 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Serial crystallography captures dynamic control of sequential electron and proton transfer events in a flavoenzyme.
Nat.Chem., 14, 2022
|
|
8XTV
 
 | | Crystal structure of AaHPPD-Y14150 complex | | Descriptor: | 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION, [3-ethyl-1-methyl-2,2-bis(oxidanylidene)-2$l^{6},1,3-benzothiadiazol-5-yl]-(1-methyl-5-oxidanyl-pyrazol-4-yl)methanone | | Authors: | Dong, J, Lin, H.-Y, Yang, G.-F. | | Deposit date: | 2024-01-11 | | Release date: | 2025-01-15 | | Last modified: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (1.994 Å) | | Cite: | Repurposing 4-Hydroxyphenylpyruvate dioxygenase inhibitors as novel agents for mosquito control: A structure-based design approach.
Int.J.Biol.Macromol., 315, 2025
|
|
2FGB
 
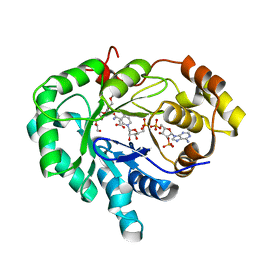 | | Crystal structure of human 17bet a-hydroxysteroid dehydrogenase type 5 in complexes with PEG and NADP | | Descriptor: | ACETATE ION, Aldo-keto reductase family 1 member C3, HEXAETHYLENE GLYCOL, ... | | Authors: | Qiu, W, Zhou, M, Azzi, A, Luu-The, V, Labrie, F, Lin, S.X. | | Deposit date: | 2005-12-21 | | Release date: | 2006-12-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure-based inhibitor design for an enzyme that binds different steroids: a potent inhibitor for human type 5 17beta-hydroxysteroid dehydrogenase.
J.Biol.Chem., 282, 2007
|
|
8XVW
 
 | | Crystal structure of N-terminal deletion mutant of Staphylococcal Thiol Peroxidase | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, GLYCEROL, SULFATE ION, ... | | Authors: | Shukla, M, Maji, S, Yadav, V.K, Das, A.K, Bhattacharyya, S. | | Deposit date: | 2024-01-15 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of N-terminal deletion mutant of Staphylococcal Thiol Peroxidase
To Be Published
|
|
6JBO
 
 | | Crystal structure of EfeO-like protein Algp7 containing samarium ion | | Descriptor: | 1,2-ETHANEDIOL, Alginate-binding protein, CITRIC ACID | | Authors: | Okumura, K, Takase, R, Maruyama, Y, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2019-01-26 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Rare metal binding by a cell-surface component of bacterial EfeUOB iron importer
To Be Published
|
|
2FGZ
 
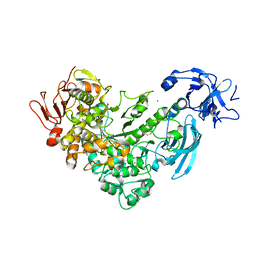 | | Crystal Structure Analysis of apo pullulanase from Klebsiella pneumoniae | | Descriptor: | Alpha-dextrin endo-1,6-alpha-glucosidase, CALCIUM ION | | Authors: | Mikami, B, Iwamoto, H, Katsuya, Y, Yoon, H.-J, Demirkan-Sarikaya, E, Malle, D. | | Deposit date: | 2005-12-23 | | Release date: | 2006-06-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of pullulanase: evidence for parallel binding of oligosaccharides in the active site
J.Mol.Biol., 359, 2006
|
|
8YJI
 
 | |
6CFO
 
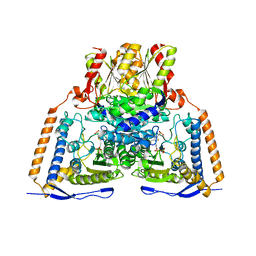 | | HUMAN PYRUVATE DEHYDROGENASE E1 COMPONENT COMPLEX WITH COVALENT TDP ADDUCT ACETYL PHOSPHINATE | | Descriptor: | 3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-2-{(1S)-1-hydroxy-1-[(R)-hydroxy(oxo)-lambda~5~-phosphanyl]ethyl}-5-(2-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-4-methyl-1,3-thiazol-3-ium, MAGNESIUM ION, Pyruvate dehydrogenase E1 component subunit alpha, ... | | Authors: | ARJUNAN, P, WHITLEY, M.J, FUREY, W. | | Deposit date: | 2018-02-15 | | Release date: | 2018-07-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Pyruvate dehydrogenase complex deficiency is linked to regulatory loop disorder in the alpha V138M variant of human pyruvate dehydrogenase.
J. Biol. Chem., 293, 2018
|
|
8YJJ
 
 | |
6CUR
 
 | | Ras:SOS:Ras in complex with a small molecule activator | | Descriptor: | FORMIC ACID, GLYCEROL, GTPase HRas, ... | | Authors: | Phan, J, Abbott, J, Fesik, S.W. | | Deposit date: | 2018-03-26 | | Release date: | 2019-02-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Discovery of Quinazolines That Activate SOS1-Mediated Nucleotide Exchange on RAS.
ACS Med Chem Lett, 9, 2018
|
|
6URA
 
 | | Crystal structure of RUBISCO from Promineofilum breve | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, Ribulose bisphosphate carboxylase large chain | | Authors: | Pereira, J.H, Banda, D.M, Liu, A.K, Shih, P.M, Adams, P.D. | | Deposit date: | 2019-10-23 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Novel bacterial clade reveals origin of form I Rubisco.
Nat.Plants, 6, 2020
|
|
9HH4
 
 | | Crystal structure of the family S1_19 carrageenan sulfatase ZgCgsA from Zobellia galactanivorans in complex with hybrid b-k-neocarratetraose | | Descriptor: | 3,6-anhydro-alpha-D-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-3,6-anhydro-alpha-D-galactopyranose-(1-3)-4-O-sulfo-beta-D-galactopyranose, BROMIDE ION, CALCIUM ION, ... | | Authors: | Chevenier, A, Czjzek, M, Michel, G, Ficko-Blean, E. | | Deposit date: | 2024-11-21 | | Release date: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure, function and catalytic mechanism of the carrageenan-sulfatases from the marine bacterium Zobellia galactanivorans Dsij T.
Carbohydr Polym, 358, 2025
|
|
6IYF
 
 | | Structure of pSTING complex | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, SULFATE ION, Stimulator of interferon genes protein | | Authors: | Yuan, Z.L, Shang, G.J, Cong, X.Y, Gu, L.C. | | Deposit date: | 2018-12-15 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.764 Å) | | Cite: | Crystal structures of porcine STINGCBD-CDN complexes reveal the mechanism of ligand recognition and discrimination of STING proteins.
J.Biol.Chem., 294, 2019
|
|
9IHV
 
 | |
8XR5
 
 | | Crystal structure of PD-L1 complexed with small molecule inhibitor X18 | | Descriptor: | (2~{R})-2-[[2-(2,1,3-benzoxadiazol-5-ylmethoxy)-5-chloranyl-4-[[2-fluoranyl-3-[3-[3-(4-oxidanylpiperidin-1-yl)propoxy]phenyl]phenyl]methoxy]phenyl]methylamino]-3-oxidanyl-propanoic acid, Programmed cell death 1 ligand 1 | | Authors: | Liu, L. | | Deposit date: | 2024-01-06 | | Release date: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of Novel PD-L1 Small-Molecular Inhibitors with Potent In Vivo Anti-tumor Immune Activity.
J.Med.Chem., 67, 2024
|
|
8X7H
 
 | | Crystal structure of the ternary complex of GID4-PROTAC(NEP162)-BRD4(BD1). | | Descriptor: | Bromodomain-containing protein 4, GLYCEROL, Glucose-induced degradation protein 4 homolog, ... | | Authors: | Dong, C, Yan, X, Li, Y. | | Deposit date: | 2023-11-24 | | Release date: | 2025-03-26 | | Last modified: | 2025-10-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Design of PROTACs utilizing the E3 ligase GID4 for targeted protein degradation.
Nat.Struct.Mol.Biol., 32, 2025
|
|
2FHB
 
 | | Crystal Structure Analysis of Klebsiella pneumoniae pullulanase complexed with maltose | | Descriptor: | Alpha-dextrin endo-1,6-alpha-glucosidase, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Mikami, B, Iwamoto, H, Katsuya, Y, Yoon, H.-J, Demirkan-Sarikaya, E, Malle, D. | | Deposit date: | 2005-12-23 | | Release date: | 2006-06-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of pullulanase: evidence for parallel binding of oligosaccharides in the active site
J.Mol.Biol., 359, 2006
|
|
8PNN
 
 | | 80S yeast ribosome in complex with Bromolissoclimide | | Descriptor: | (3~{R})-3-[(1~{S})-2-[(1~{R},3~{S},4~{a}~{S},7~{S},8~{a}~{S})-7-bromanyl-5,5,8~{a}-trimethyl-2-methylidene-3-oxidanyl-3,4,4~{a},6,7,8-hexahydro-1~{H}-naphthalen-1-yl]-1-oxidanyl-ethyl]pyrrolidine-2,5-dione, 16S ribosomal RNA, 25S ribosomal RNA, ... | | Authors: | Terrosu, S, Yusupov, M. | | Deposit date: | 2023-06-30 | | Release date: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Synthesis of Differentially Halogenated Lissoclimide Analogues To Probe Ribosome E-Site Binding.
Acs Chem.Biol., 20, 2025
|
|
6J3F
 
 | | Crystal structure of the glutathione S-transferase, CsGST63524, of Ceriporiopsis subvermispora in complex with glutathione | | Descriptor: | 1,2-ETHANEDIOL, GLUTATHIONE, glutathione S-transferase | | Authors: | Osman, W.H.W, Mikami, B, Saka, N, Kondo, K, Nagata, T, Katahira, M. | | Deposit date: | 2019-01-04 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a serine-type glutathione S-transferase of Ceriporiopsis subvermispora and identification of the enzymatically important non-canonical residues by functional mutagenesis.
Biochem. Biophys. Res. Commun., 510, 2019
|
|
8Y6C
 
 | | Norovirus GII.10 P domain and 2'-FL (powder) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GII.10 norovirus P domain in complex with 2'FL (powder), ... | | Authors: | Hansman, G, Tame, J.R.H, Kher, G, Pancera, M. | | Deposit date: | 2024-02-02 | | Release date: | 2024-06-19 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Human milk oligosaccharide 2'-fucosyllactose guards norovirus histo-blood group antigen co-factor binding site.
J.Virol., 98, 2024
|
|
8Y6D
 
 | | Norovirus GII.10 P domain and 2'-FL (tablet) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GII.10 norovirus P domain in complex with 2'-FL (tablet), ... | | Authors: | Hansman, G, Tame, J.R.H, Kher, G, Pancera, M. | | Deposit date: | 2024-02-02 | | Release date: | 2024-06-19 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Human milk oligosaccharide 2'-fucosyllactose guards norovirus histo-blood group antigen co-factor binding site.
J.Virol., 98, 2024
|
|
6J3E
 
 | | Crystal structure of an apo form of the glutathione S-transferase, CsGST63524, of Ceriporiopsis subvermispora | | Descriptor: | 1,2-ETHANEDIOL, glutathione S-transferase | | Authors: | Osman, W.H.W, Mikami, B, Saka, N, Kondo, K, Nagata, T, Katahira, M. | | Deposit date: | 2019-01-04 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.455 Å) | | Cite: | Structure of a serine-type glutathione S-transferase of Ceriporiopsis subvermispora and identification of the enzymatically important non-canonical residues by functional mutagenesis.
Biochem. Biophys. Res. Commun., 510, 2019
|
|
