1IA3
 
 | | Candida albicans dihydrofolate reductase complex in which the dihydronicotinamide moiety of dihydro-nicotinamide-adenine-dinucleotide phosphate (NADPH) is displaced by 5-[(4-TERT-BUTYLPHENYL)SULFANYL]-2,4-QUINAZOLINEDIAMINE (GW995) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-[4-TERT-BUTYLPHENYLSULFANYL]-2,4-QUINAZOLINEDIAMINE, DIHYDROFOLATE REDUCTASE, ... | | Authors: | Whitlow, M, Howard, A.J, Kuyper, L.F. | | Deposit date: | 2001-03-22 | | Release date: | 2001-04-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | X-ray Crystal Structures of Candida albicans Dihydrofolate Reductase: High Resolution Ternary Complexes in Which the Dihydronicotinamide Moiety of NADPH is Displaced by an inhibitor
J.Med.Chem., 44, 2001
|
|
5GVC
 
 | | Human Topoisomerase IIIb topo domain | | Descriptor: | DNA topoisomerase 3-beta-1, MAGNESIUM ION | | Authors: | Goto-Ito, S, Yamagata, A, Sato, Y, Takahashi, T.S, Fukai, S. | | Deposit date: | 2016-09-05 | | Release date: | 2017-06-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.436 Å) | | Cite: | Structural basis of the interaction between Topoisomerase III beta and the TDRD3 auxiliary factor
Sci Rep, 7, 2017
|
|
1IA4
 
 | | Candida albicans dihydrofolate reductase complex in which the dihydronicotinamide moiety of dihydro-nicotinamide-adenine-dinucleotide phosphate (NADPH) is displaced by 5-{[4-(4-MORPHOLINYL)PHENYL]SULFANYL}-2,4-QUINAZOLINEDIAMIN (GW2021) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-(4-MORPHOLIN-4-YL-PHENYLSULFANYL)-2,4-QUINAZOLINEDIAMINE, DIHYDROFOLATE REDUCTASE, ... | | Authors: | Whitlow, M, Howard, A.J, Kuyper, L.F. | | Deposit date: | 2001-03-22 | | Release date: | 2001-04-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-ray Crystal Structures of Candida albicans Dihydrofolate Reductase: High Resolution Ternary Complexes in Which the Dihydronicotinamide Moiety of NADPH is Displaced by an inhibitor
J.Med.Chem., 44, 2001
|
|
5F9O
 
 | | Crystal structure of broadly neutralizing VH1-46 germline-derived CD4-binding site-directed antibody CH235.09 in complex with HIV-1 clade A/E 93TH057 gp120 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CH235.09 Light chain, ... | | Authors: | Joyce, M.G, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2015-12-10 | | Release date: | 2016-03-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Maturation Pathway from Germline to Broad HIV-1 Neutralizer of a CD4-Mimic Antibody.
Cell, 165, 2016
|
|
5FA0
 
 | | The structure of the beta-3-deoxy-D-manno-oct-2-ulosonic acid transferase domain from WbbB | | Descriptor: | CHLORIDE ION, Putative N-acetyl glucosaminyl transferase | | Authors: | Mallette, E, Ovchinnikova, O.G, Whitfield, C, Kimber, M.S. | | Deposit date: | 2015-12-10 | | Release date: | 2016-05-18 | | Last modified: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bacterial beta-Kdo glycosyltransferases represent a new glycosyltransferase family (GT99).
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
2YPQ
 
 | | 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase with tryptophan and tyrosine bound | | Descriptor: | CHLORIDE ION, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Blackmore, N.J, Reichau, S, Jiao, W, Hutton, R.D, Baker, E.N, Jameson, G.B, Parker, E.J. | | Deposit date: | 2012-10-31 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Three Sites and You are Out: Ternary Synergistic Allostery Controls Aromatic Aminoacid Biosynthesis in Mycobacterium Tuberculosis.
J.Mol.Biol., 425, 2013
|
|
3X38
 
 | | Crystal structure of the C-terminal domain of Sld7 | | Descriptor: | GLYCEROL, Mitochondrial morphogenesis protein SLD7, SULFATE ION | | Authors: | Itou, H, Araki, H, Shirakihara, Y. | | Deposit date: | 2015-01-16 | | Release date: | 2015-08-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | The quaternary structure of the eukaryotic DNA replication proteins Sld7 and Sld3.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1IA1
 
 | | Candida albicans dihydrofolate reductase complexed with dihydro-nicotinamide-adenine-dinucleotide phosphate (NADPH) and 5-(PHENYLSULFANYL)-2,4-QUINAZOLINEDIAMINE (GW997) | | Descriptor: | 5-PHENYLSULFANYL-2,4-QUINAZOLINEDIAMINE, DIHYDROFOLATE REDUCTASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Whitlow, M, Howard, A.J, Kuyper, L.F. | | Deposit date: | 2001-03-22 | | Release date: | 2001-04-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray Crystal Structures of Candida albicans Dihydrofolate Reductase: High Resolution Ternary Complexes in Which the Dihydronicotinamide Moiety of NADPH is Displaced by an inhibitor
J.Med.Chem., 44, 2001
|
|
1IA2
 
 | | Candida albicans dihydrofolate reductase complexed with dihydro-nicotinamide-adenine-dinucleotide phosphate (NADPH) and 5-[(4-METHYLPHENYL)SULFANYL]-2,4-QUINAZOLINEDIAMINE (GW578) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-[(4-METHYLPHENYL)SULFANYL]-2,4-QUINAZOLINEDIAMINE, DIHYDROFOLATE REDUCTASE, ... | | Authors: | Whitlow, M, Howard, A.J, Kuyper, L.F. | | Deposit date: | 2001-03-22 | | Release date: | 2001-04-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | X-ray Crystal Structures of Candida albicans Dihydrofolate Reductase: High Resolution Ternary Complexes in Which the Dihydronicotinamide Moiety of NADPH is Displaced by an inhibitor
J.Med.Chem., 44, 2001
|
|
3EPJ
 
 | |
3TL4
 
 | |
3EPK
 
 | |
8WCQ
 
 | | Cryo-EM structure of nanodisc (PE:PS:PC) reconstituted GLIC at pH 4 in intermediate state | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CHLORIDE ION, Proton-gated ion channel | | Authors: | Bharambe, N, Li, Z, Basak, S. | | Deposit date: | 2023-09-13 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Cryo-EM structures of prokaryotic ligand-gated ion channel GLIC provide insights into gating in a lipid environment.
Nat Commun, 15, 2024
|
|
9CEL
 
 | | Juvenimicin Thioesterase | | Descriptor: | Type I PKS module 7 | | Authors: | Akey, D.L, Smith, J.S, Choudhary, V. | | Deposit date: | 2024-06-26 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Substrate Trapping in Polyketide Synthase Thioesterase Domains: Structural Basis for Macrolactone Formation
Acs Catalysis, 14, 2024
|
|
8WCR
 
 | | Cryo-EM structure of nanodisc (PE:PS:PC) reconstituted GLIC at pH 4 in open state | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CHLORIDE ION, Proton-gated ion channel | | Authors: | Bharambe, N, Li, Z, Basak, S. | | Deposit date: | 2023-09-13 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Cryo-EM structures of prokaryotic ligand-gated ion channel GLIC provide insights into gating in a lipid environment.
Nat Commun, 15, 2024
|
|
9CGO
 
 | | Tylosin thioesterase domain (TylG5 TE) | | Descriptor: | Tylactone synthase module 7 | | Authors: | Smith, J.L, Choudhary, V. | | Deposit date: | 2024-06-30 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Substrate Trapping in Polyketide Synthase Thioesterase Domains: Structural Basis for Macrolactone Formation
Acs Catalysis, 14, 2024
|
|
8YF5
 
 | | Cryo EM structure of Komagataella phaffii Rat1-Rai1-Rtt103 complex | | Descriptor: | 5'-3' exoribonuclease, Decapping nuclease, Exonuclease Rat1p and Rai1p interacting protein | | Authors: | Yanagisawa, T, Murayama, Y, Ehara, H, Sekine, S.I. | | Deposit date: | 2024-02-24 | | Release date: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Structural basis of eukaryotic transcription termination by the Rat1 exonuclease complex.
Nat Commun, 15, 2024
|
|
9G7H
 
 | | Human Sirt6 in complex with ADP-ribose and the inhibitor 2-Pr | | Descriptor: | 2,4-bis(oxidanylidene)-1-[2-oxidanylidene-2-[[(2S)-3-oxidanylidene-3-(propylamino)-2-[3-(3-pyridin-3-yl-1,2,4-oxadiazol-5-yl)propanoylamino]propyl]amino]ethyl]pyrimidine-5-carboxamide, NAD-dependent protein deacetylase sirtuin-6, SULFATE ION, ... | | Authors: | You, W, Steegborn, C. | | Deposit date: | 2024-07-21 | | Release date: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Elucidating the Unconventional Binding Mode of a DNA-Encoded Library Hit Provides a Blueprint for Sirtuin 6 Inhibitor Development.
Chemmedchem, 2024
|
|
8YFE
 
 | | Cryo EM structure of Komagataella phaffii Rat1-Rai1 complex | | Descriptor: | 5'-3' exoribonuclease, Decapping nuclease | | Authors: | Yanagisawa, T, Murayama, Y, Ehara, H, Sekine, S.I. | | Deposit date: | 2024-02-24 | | Release date: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structural basis of eukaryotic transcription termination by the Rat1 exonuclease complex.
Nat Commun, 15, 2024
|
|
8W0K
 
 | |
8VRE
 
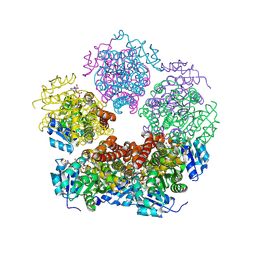 | | Structure of PYCR1 complexed with NADH and N-formyl-L-proline | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 1-formyl-L-proline, Pyrroline-5-carboxylate reductase 1, ... | | Authors: | Tanner, J.J, Meeks, K.R. | | Deposit date: | 2024-01-21 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Screening a knowledge-based library of low molecular weight compounds against the proline biosynthetic enzyme 1-pyrroline-5-carboxylate 1 (PYCR1).
Protein Sci., 33, 2024
|
|
8X3N
 
 | |
8X40
 
 | |
9BOR
 
 | | IkappaBzeta ankyrin repeat domain:NF-kappaB p50 homodimer complex at 2.0 Angstrom resolution | | Descriptor: | (2R,3S)-heptane-1,2,3-triol, NF-kappa-B inhibitor zeta, Nuclear factor NF-kappa-B p50 subunit | | Authors: | Rogers, W.E, Zhu, N, Huxford, T. | | Deposit date: | 2024-05-05 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Structural and biochemical analyses of the nuclear I kappa B zeta protein in complex with the NF-kappa B p50 homodimer.
Genes Dev., 38, 2024
|
|
2VRL
 
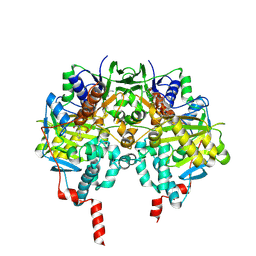 | | Structure of human MAO B in complex with benzylhydrazine | | Descriptor: | AMINE OXIDASE [FLAVIN-CONTAINING] B, FLAVIN-ADENINE DINUCLEOTIDE, TOLUENE | | Authors: | Binda, C, Wang, J, Li, M, Hubalek, F, Mattevi, A, Edmondson, D.E. | | Deposit date: | 2008-04-09 | | Release date: | 2008-04-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Mechanistic Studies of Arylalkylhydrazine Inhibition of Human Monoamine Oxidases a and B
Biochemistry, 47, 2008
|
|
