3FKG
 
 | | Crystal Structure Analysis of Fungal Versatile Peroxidase from Pleurotus eryngii | | Descriptor: | CACODYLATE ION, CALCIUM ION, FE (III) ION, ... | | Authors: | Piontek, K, Martinez, A.T, Choinowski, T, Plattner, D.A. | | Deposit date: | 2008-12-16 | | Release date: | 2009-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural and Site-directed Mutagenesis Study of Versatile Peroxidase Oxidizing both Mn(II) and Aromatic Substrates
To be Published
|
|
3F29
 
 | | Structure of the Thioalkalivibrio nitratireducens cytochrome c nitrite reductase in complex with sulfite | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, CALCIUM ION, Eight-heme nitrite reductase, ... | | Authors: | Trofimov, A.A, Polyakov, K.M, Boyko, K.M, Slutsky, A, Tikhonova, T.V, Antipov, A.N, Zvyagilskaya, R.A, Popov, A.N, Lamzin, V.S, Bourenkov, G.P, Popov, V.O. | | Deposit date: | 2008-10-29 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding of sulfite by the Thioalkalivibrio nitratireducens cytochrome c nitrite reductase
To be Published
|
|
3FLU
 
 | |
2VCV
 
 | |
2VJX
 
 | | Structural and biochemical evidence for a boat-like transition state in beta-mannosidases | | Descriptor: | (3S,4R,5R)-3,4-DIHYDROXY-5-(HYDROXYMETHYL)PIPERIDIN-2-ONE, 1,2-ETHANEDIOL, BETA-MANNOSIDASE, ... | | Authors: | Tailford, L.E, Offen, W.A, Smith, N.L, Dumon, C, Moreland, C, Gratien, J, Heck, M.P, Stick, R.V, Bleriot, Y, Vasella, A, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2007-12-14 | | Release date: | 2008-04-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Biochemical Evidence for a Boat-Like Transition State in Beta-Mannosidases.
Nat.Chem.Biol., 4, 2008
|
|
2VI5
 
 | | LUMAZINE SYNTHASE FROM MYCOBACTERIUM TUBERCULOSIS BOUND TO N-6-(ribitylamino)pyrimidine-2,4(1H,3H)-dione-5-yl-propionamide | | Descriptor: | 1-deoxy-1-{[(5S)-2,6-dioxo-5-(propanoylamino)-1,2,5,6-tetrahydropyrimidin-4-yl]amino}-D-ribitol, 6,7-DIMETHYL-8-RIBITYLLUMAZINE SYNTHASE, PHOSPHATE ION, ... | | Authors: | Morgunova, E, Zhang, Y, Jin, G, Illarionov, B, Bacher, A, Fischer, M, Cushman, M, Ladenstein, R. | | Deposit date: | 2007-11-27 | | Release date: | 2008-04-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A New Series of N-[2,4-Dioxo-6-D-Ribitylamino-1,2, 3,4-Tetrahydropyrimidin-5-Yl]Oxalamic Acid Derivatives as Inhibitors of Lumazine Syntase and Riboflavin Synthase: Design, Synthesis, Biochemical Evaluation, Crystallography and Mechanistic Implications.
J.Org.Chem., 73, 2008
|
|
3E8H
 
 | |
2UZJ
 
 | | Crystal structure of the mature streptococcal cysteine protease, mSpeB | | Descriptor: | N-[N-[1-HYDROXYCARBOXYETHYL-CARBONYL]LEUCYLAMINO-BUTYL]-GUANIDINE, STREPTOPAIN | | Authors: | Olsen, J.G, Dagil, R, Niclasen, L.M, Soerensen, O.E, Kragelund, B.B. | | Deposit date: | 2008-09-16 | | Release date: | 2009-09-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of the Mature Streptococcal Cysteine Protease Exotoxin Mspeb in its Active Dimeric Form.
J.Mol.Biol., 393, 2009
|
|
3RTA
 
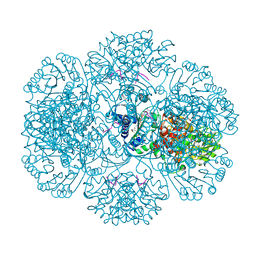 | | Crystal structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima soaked with Acetyl Coenzyme A | | Descriptor: | ACETYL COENZYME *A, POTASSIUM ION, Putative uncharacterized protein, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | Deposit date: | 2011-05-03 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3R0H
 
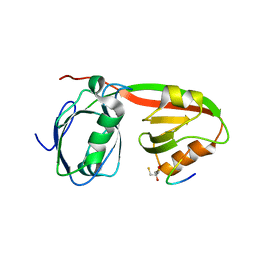 | | Structure of INAD PDZ45 in complex with NG2 peptide | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Inactivation-no-after-potential D protein, ... | | Authors: | Wei, Z, Liu, W, Zhang, M. | | Deposit date: | 2011-03-08 | | Release date: | 2011-11-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The INAD scaffold is a dynamic, redox-regulated modulator of signaling in the Drosophila eye
Cell(Cambridge,Mass.), 145, 2011
|
|
2V19
 
 | |
3UJJ
 
 | |
3UK9
 
 | | Galactose-specific lectin from Dolichos lablab | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Shetty, K.N, Latha, V.L, Rao, R.N, Nadimpalli, S.K, Suguna, K. | | Deposit date: | 2011-11-09 | | Release date: | 2012-11-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Affinity of a galactose-specific legume lectin from Dolichos lablab to adenine revealed by X-ray cystallography.
Iubmb Life, 65, 2013
|
|
2PLS
 
 | | Structural Genomics, the crystal structure of the CorC/HlyC transporter associated domain of a CBS domain protein from Chlorobium tepidum TLS | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CBS domain protein, ... | | Authors: | Tan, K, Volkart, L, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-04-20 | | Release date: | 2007-05-22 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The crystal structure of the CorC/HlyC transporter associated domain of a CBS domain protein from Chlorobium tepidum TLS.
To be Published
|
|
3UNO
 
 | |
3RGD
 
 | | Iron loaded frog M ferritin. Short soaking time | | Descriptor: | FE (III) ION, Ferritin, middle subunit | | Authors: | Bertini, I, Lalli, D, Mangani, S, Pozzi, C, Rosa, C, Theil, E.C, Turano, P. | | Deposit date: | 2011-04-08 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structural insights into the ferroxidase site of ferritins from higher eukaryotes.
J.Am.Chem.Soc., 134, 2012
|
|
3IR0
 
 | |
3REL
 
 | |
3UT9
 
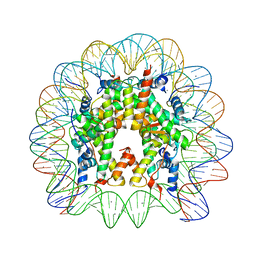 | | Crystal Structure of Nucleosome Core Particle Assembled with a Palindromic Widom '601' Derivative (NCP-601L) | | Descriptor: | 145-mer DNA, CHLORIDE ION, Histone H2A, ... | | Authors: | Chua, E.Y.D, Vasudevan, D, Davey, G.E, Wu, B, Davey, C.A. | | Deposit date: | 2011-11-25 | | Release date: | 2012-04-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The mechanics behind DNA sequence-dependent properties of the nucleosome
Nucleic Acids Res., 40, 2012
|
|
3UTA
 
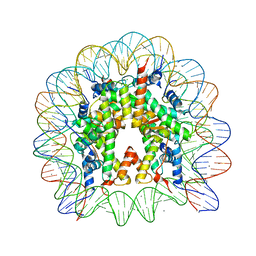 | | Crystal Structure of Nucleosome Core Particle Assembled with an Alpha-Satellite Sequence Containing Two TTAAA elements (NCP-TA2) | | Descriptor: | 145-mer DNA, CHLORIDE ION, Histone H2A, ... | | Authors: | Chua, E.Y.D, Vasudevan, D, Davey, G.E, Wu, B, Davey, C.A. | | Deposit date: | 2011-11-25 | | Release date: | 2012-04-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | The mechanics behind DNA sequence-dependent properties of the nucleosome
Nucleic Acids Res., 40, 2012
|
|
3E4F
 
 | | Crystal structure of BA2930- a putative aminoglycoside N3-acetyltransferase from Bacillus anthracis | | Descriptor: | Aminoglycoside N3-acetyltransferase, CITRIC ACID | | Authors: | Klimecka, M.M, Chruszcz, M, Skarina, T, Onopryienko, O, Cymborowski, M, Savchenko, A, Edwards, A, Anderson, W, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-08-11 | | Release date: | 2008-08-19 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analysis of a Putative Aminoglycoside N-Acetyltransferase from Bacillus anthracis.
J.Mol.Biol., 410, 2011
|
|
3I9V
 
 | | Crystal structure of the hydrophilic domain of respiratory complex I from Thermus thermophilus, oxidized, 2 mol/ASU | | Descriptor: | CALCIUM ION, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Sazanov, L.A, Berrisford, J.M. | | Deposit date: | 2009-07-13 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for the mechanism of respiratory complex I
J.Biol.Chem., 284, 2009
|
|
3IAI
 
 | | Crystal structure of the catalytic domain of the tumor-associated human carbonic anhydrase IX | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, Carbonic anhydrase 9, ... | | Authors: | Alterio, V, Di Fiore, A, De Simone, G. | | Deposit date: | 2009-07-14 | | Release date: | 2009-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the catalytic domain of the tumor-associated human carbonic anhydrase IX.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3ROJ
 
 | | D-fructose 1,6-bisphosphatase class 2/sedoheptulose 1,7-bisphosphatase of Synechocystis sp. PCC 6803 | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, D-fructose 1,6-bisphosphatase class 2/sedoheptulose 1,7-bisphosphatase, ... | | Authors: | Hu, X, Hui, D, Lingling, F, Jian, W. | | Deposit date: | 2011-04-26 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | New insights into the structural and interactional basis for a promising route towards fructose-1,6-/sedoheptulose-1,7-bisphosphatases controlling
To be Published
|
|
3IDY
 
 | | Crystal structure of HIV-gp120 core in complex with CD4-binding site antibody b13, space group C2221 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab b13 heavy chain, Fab b13 light chain, ... | | Authors: | Chen, L, Kwon, Y.D, Zhou, T, Wu, X, O'Dell, S, Cavacini, L, Hessell, A.J, Pancera, M, Tang, M, Xu, L, Yang, Z.Y, Zhang, M.Y, Arthos, J, Burton, D.R, Dimitrov, D.S, Nabel, G.J, Posner, M, Sodroski, J, Wyatt, R, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2009-07-22 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120.
Science, 326, 2009
|
|
