8DNF
 
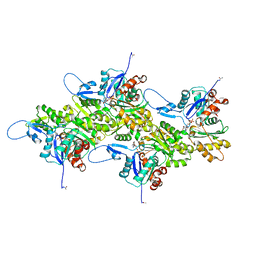 | | Cryo-EM structure of nonmuscle gamma-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 2, ... | | Authors: | Arora, A.S, Huang, H.L, Heissler, S.M, Chinthalapudi, K. | | Deposit date: | 2022-07-11 | | Release date: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structural insights into actin isoforms.
Elife, 12, 2023
|
|
8DNH
 
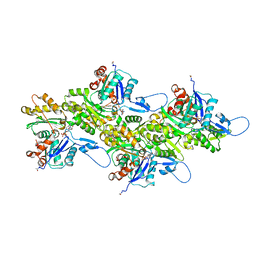 | | Cryo-EM structure of nonmuscle beta-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Arora, A.S, Huang, H.L, Heissler, S.M, Chinthalapudi, K. | | Deposit date: | 2022-07-11 | | Release date: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Structural insights into actin isoforms.
Elife, 12, 2023
|
|
8DND
 
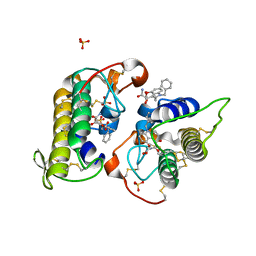 | |
8ADT
 
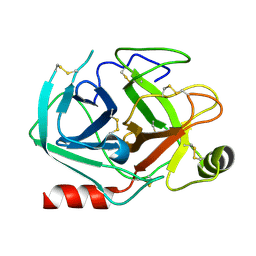 | | Rational design of a calcium-independent trypsin variant | | Descriptor: | Serine protease 1 | | Authors: | Simon, A.H, Liebscher, S, Kattner, A, Kattner, C, Bordusa, F. | | Deposit date: | 2022-07-11 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Rational Design of a Calcium-Independent Trypsin Variant
Catalysts, 12, 2022
|
|
7YG5
 
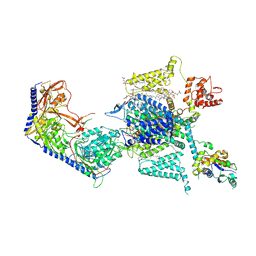 | |
8DNC
 
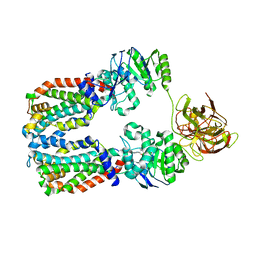 | | CryoEM structure of the A. aeolicus WzmWzt transporter bound to the native O antigen and ADP | | Descriptor: | 6-deoxy-3-O-methyl-alpha-D-mannopyranose-(1-3)-beta-D-rhamnopyranose-(1-2)-alpha-D-rhamnopyranose, ABC transporter, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Spellmon, N, Muszynski, A, Vlach, J, Zimmer, J. | | Deposit date: | 2022-07-11 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis for polysaccharide recognition and modulated ATP hydrolysis by the O antigen ABC transporter.
Nat Commun, 13, 2022
|
|
8DNE
 
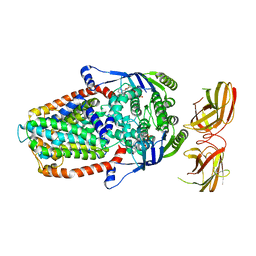 | |
8DNS
 
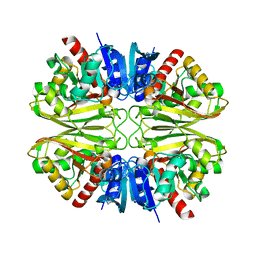 | | Human Brain Glyceraldehyde 3-phosphate dehydrogenase | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase | | Authors: | Tringides, M.L. | | Deposit date: | 2022-07-11 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | A cryo-electron microscopic approach to elucidate protein structures from human brain microsomes.
Life Sci Alliance, 6, 2023
|
|
8ADR
 
 | |
8DN1
 
 | | Q108K:K40L:T51C:T53A:R58L:Q38F:Q4F mutant of hCRBPII bound to synthetic fluorophore CM1V at pH 7.2 | | Descriptor: | (2E)-3-[7-(diethylamino)-2-oxo-2H-1-benzopyran-3-yl]prop-2-enal, bound form, GLYCEROL, ... | | Authors: | Bingham, C.R, Borhan, B, Geiger, J.H. | | Deposit date: | 2022-07-10 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Light controlled reversible Michael addition of cysteine: a new tool for dynamic site-specific labeling of proteins.
Analyst, 148, 2023
|
|
8DN8
 
 | |
8DMY
 
 | | Cryo-EM structure of cardiac muscle alpha-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha cardiac muscle 1, ... | | Authors: | Arora, A.S, Huang, H.L, Heissler, S.M, Chinthalapudi, K. | | Deposit date: | 2022-07-09 | | Release date: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural insights into actin isoforms.
Elife, 12, 2023
|
|
7YFR
 
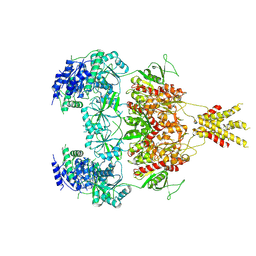 | | Structure of GluN1a E698C-GluN2D NMDA receptor in cystines non-crosslinked state. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Zhang, J.L, Zhu, S.J, Zhang, M. | | Deposit date: | 2022-07-09 | | Release date: | 2023-04-12 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7YFT
 
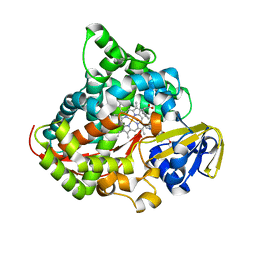 | | Crystal structure of the P450 BM3 heme domain mutant F87A/T268V/A82C/L181M in complex with N-imidazolyl-pentanoyl-L-phenylalanine, indane and hydroxylamine | | Descriptor: | (2~{S})-2-(5-imidazol-1-ylpentanoylamino)-3-phenyl-propanoic acid, 2,3-dihydro-1H-indene, Bifunctional cytochrome P450/NADPH--P450 reductase, ... | | Authors: | Dong, S, Chen, J, Jiang, Y, Cong, Z, Feng, Y. | | Deposit date: | 2022-07-09 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Regiodivergent and Enantioselective Hydroxylation of C-H bonds by Synergistic Use of Protein Engineering and Exogenous Dual-Functional Small Molecules.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8DMO
 
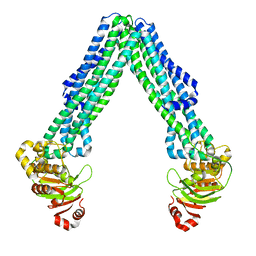 | | Structure of open, inward-facing MsbA from E. coli | | Descriptor: | ATP-binding transport protein MsbA | | Authors: | Liu, C, Lyu, J, Laganowsky, A.D, Zhao, M. | | Deposit date: | 2022-07-08 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for lipid and copper regulation of the ABC transporter MsbA.
Nat Commun, 13, 2022
|
|
7YFI
 
 | | Structure of the Rat tri-heteromeric GluN1-GluN2A-GluN2C NMDA receptor in complex with glycine and glutamate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Zhang, M, Zhang, J, Guo, F, Li, Y, Zhu, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-03-29 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7YFC
 
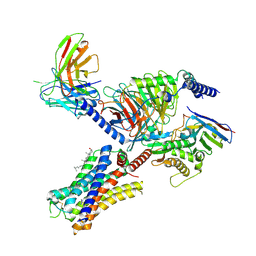 | | Cryo-EM structure of the histamine-bound histamine H4 receptor and Gq complex | | Descriptor: | CHOLESTEROL, Engineered G-alpha-q, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Im, D, Iwata, S, Asada, H. | | Deposit date: | 2022-07-08 | | Release date: | 2023-10-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the agonists binding and receptor selectivity of human histamine H 4 receptor.
Nat Commun, 14, 2023
|
|
7YFD
 
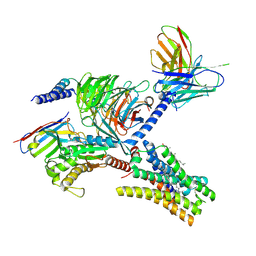 | | Cryo-EM structure of the imetit-bound histamine H4 receptor and Gq complex | | Descriptor: | 2-(1~{H}-imidazol-5-yl)ethyl carbamimidothioate, CHOLESTEROL, Engineered G-alpha-q, ... | | Authors: | Im, D, Iwata, S, Asada, H. | | Deposit date: | 2022-07-08 | | Release date: | 2023-10-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the agonists binding and receptor selectivity of human histamine H 4 receptor.
Nat Commun, 14, 2023
|
|
8DMX
 
 | | Cryo-EM structure of skeletal muscle alpha-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Arora, A.S, Huang, H.L, Heissler, S.M, Chinthalapudi, K. | | Deposit date: | 2022-07-08 | | Release date: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Structural insights into actin isoforms.
Elife, 12, 2023
|
|
7YFN
 
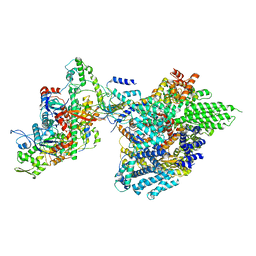 | | Core module of the NuA4 complex in S. cerevisiae | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ARP4 isoform 1, Actin, ... | | Authors: | Ji, L.T, Zhao, L.X, Xu, K, Gao, H.H, Zhou, Y, Kornberg, R.D, Zhang, H.Q. | | Deposit date: | 2022-07-08 | | Release date: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the NuA4 histone acetyltransferase complex.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7YFG
 
 | | Structure of the Rat GluN1-GluN2C NMDA receptor in complex with glycine and glutamate (major class in asymmetry) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Zhang, M, Zhang, J, Guo, F, Li, Y, Zhu, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-03-29 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7YFM
 
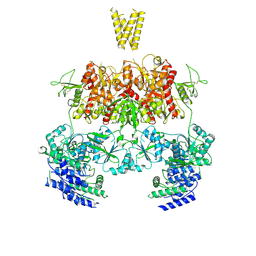 | | Structure of GluN1b-GluN2D NMDA receptor in complex with agonists glycine and glutamate. | | Descriptor: | Glutamate receptor ionotropic, NMDA 2D, Isoform 6 of Glutamate receptor ionotropic, ... | | Authors: | Zhang, J.L, Zhu, S.J, Zhang, M. | | Deposit date: | 2022-07-08 | | Release date: | 2023-03-29 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7YFH
 
 | | Structure of the Rat GluN1-GluN2C NMDA receptor in complex with glycine, glutamate and (R)-PYD-106 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Zhang, M, Zhang, J, Guo, F, Li, Y, Zhu, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-03-29 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7YFF
 
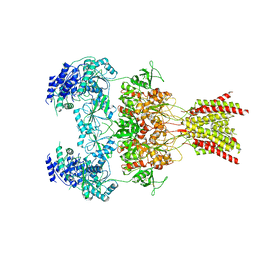 | | Structure of GluN1a-GluN2D NMDA receptor in complex with agonist glycine and competitive antagonist CPP. | | Descriptor: | (2R)-4-(3-phosphonopropyl)piperazine-2-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J.L, Zhu, S.J, Zhang, M. | | Deposit date: | 2022-07-08 | | Release date: | 2023-04-12 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7YFL
 
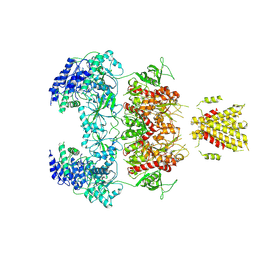 | | Structure of GluN1a-GluN2D NMDA receptor in complex with agonists glycine and glutamate. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Zhang, J.L, Zhu, S.J, Zhang, M. | | Deposit date: | 2022-07-08 | | Release date: | 2023-04-12 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
