4D3C
 
 | |
7AU7
 
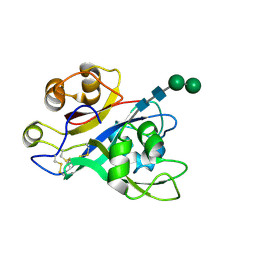 | | Crystal structure of Nod Factor Perception ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Serine/threonine receptor-like kinase NFP, ... | | Authors: | Gysel, K, Blaise, M, Andersen, K.R. | | Deposit date: | 2020-11-02 | | Release date: | 2021-11-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.547 Å) | | Cite: | Kinetic proofreading of lipochitooligosaccharides determines signal activation of symbiotic plant receptors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7AXU
 
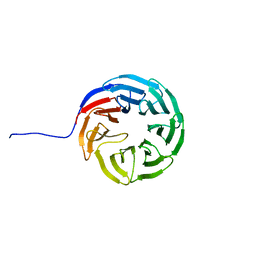 | |
7B0B
 
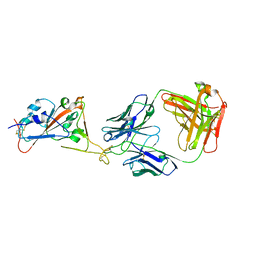 | | Fab HbnC3t1p1_C6 bound to SARS-CoV-2 RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab heavy chain, IGK@ protein, ... | | Authors: | Borenstein-Katz, A, Diskin, R. | | Deposit date: | 2020-11-19 | | Release date: | 2021-12-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Somatic hypermutation introduces bystander mutations that prepare SARS-CoV-2 antibodies for emerging variants.
Immunity, 56, 2023
|
|
3VKP
 
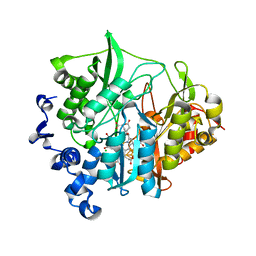 | | Assimilatory nitrite reductase (Nii3) - NO2 complex from tobbaco leaf analysed with low X-ray dose | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, NITRITE ION, ... | | Authors: | Nakano, S, Takahashi, M, Sakamoto, A, Morikawa, H, Katayanagi, K. | | Deposit date: | 2011-11-20 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The reductive reaction mechanism of tobacco nitrite reductase derived from a combination of crystal structures and ultraviolet-visible microspectroscopy
Proteins, 80, 2012
|
|
3IVY
 
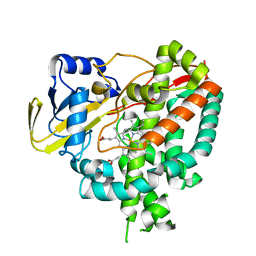 | | Crystal structure of Mycobacterium tuberculosis cytochrome P450 CYP125, p212121 crystal form | | Descriptor: | Cytochrome P450 CYP125, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | McLean, K.J, Levy, C, Munro, A.W, Leys, D. | | Deposit date: | 2009-09-02 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.352 Å) | | Cite: | The Structure of Mycobacterium tuberculosis CYP125: molecular basis for cholesterol binding in a P450 needed for host infection.
J.Biol.Chem., 284, 2009
|
|
3IW2
 
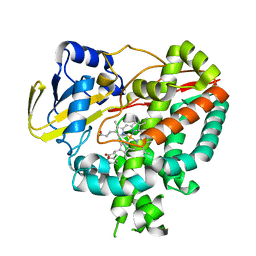 | | Crystal structure of Mycobacterium tuberculosis cytochrome P450 CYP125 in complex with econazole | | Descriptor: | 4-[(2R)-2-[(4-chlorobenzyl)oxy]-2-(2,4-dichlorophenyl)ethyl]-1H-imidazole, Cytochrome P450 CYP125, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | McLean, K.J, Levy, C, Munro, A.W, Leys, D. | | Deposit date: | 2009-09-02 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The Structure of Mycobacterium tuberculosis CYP125: molecular basis for cholesterol binding in a P450 needed for host infection.
J.Biol.Chem., 284, 2009
|
|
7QUR
 
 | | SARS-CoV-2 Spike with ethylbenzamide-tri-iodo Siallyllactose, C3 symmetry | | Descriptor: | 2,3,5-tris(iodanyl)benzamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Naismith, J.H, Yang, Y, Liu, J.W. | | Deposit date: | 2022-01-18 | | Release date: | 2022-06-01 | | Last modified: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (2.27 Å) | | Cite: | Pathogen-sugar interactions revealed by universal saturation transfer analysis.
Science, 377, 2022
|
|
7QUS
 
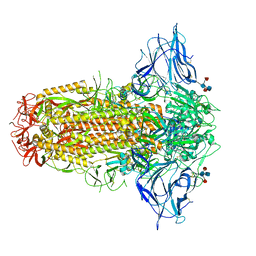 | | SARS-CoV-2 Spike, C3 symmetry | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | Authors: | Naismith, J.H, Yang, Y, Liu, J.W. | | Deposit date: | 2022-01-18 | | Release date: | 2022-06-08 | | Last modified: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (2.39 Å) | | Cite: | Pathogen-sugar interactions revealed by universal saturation transfer analysis.
Science, 377, 2022
|
|
7PAZ
 
 | | REDUCED MUTANT P80I PSEUDOAZURIN FROM A. FAECALIS | | Descriptor: | COPPER (II) ION, PSEUDOAZURIN | | Authors: | Adman, E.T, Libeu, C.A.P. | | Deposit date: | 1997-02-21 | | Release date: | 1997-08-20 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Site-directed mutants of pseudoazurin: explanation of increased redox potentials from X-ray structures and from calculation of redox potential differences.
Biochemistry, 36, 1997
|
|
7Q52
 
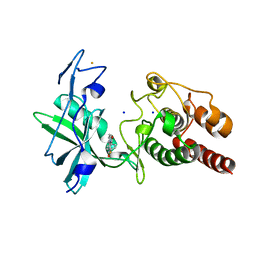 | | Crystal structure of S/T protein kinase PknG from Mycobacterium tuberculosis in complex with inhibitor L2W | | Descriptor: | 2-azanyl-3-(4-fluorophenyl)carbonyl-indolizine-1-carboxamide, FE (III) ION, SODIUM ION, ... | | Authors: | Defelipe, L.A, Burastero, O, Bento, I, Garcia-Alai, M.M. | | Deposit date: | 2021-11-02 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Cosolvent Sites-Based Discovery of Mycobacterium Tuberculosis Protein Kinase G Inhibitors.
J.Med.Chem., 65, 2022
|
|
7QXJ
 
 | |
8CX0
 
 | | Cryo-EM structure of human APOBEC3G/HIV-1 Vif/CBFbeta/ELOB/ELOC monomeric complex | | Descriptor: | Core-binding factor subunit beta, DNA dC->dU-editing enzyme APOBEC-3G, Elongin-B, ... | | Authors: | Li, Y, Langley, C, Azumaya, C.M, Echeverria, I, Chesarino, N.M, Emerman, M, Cheng, Y, Gross, J.D. | | Deposit date: | 2022-05-19 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | The structural basis for HIV-1 Vif antagonism of human APOBEC3G.
Nature, 615, 2023
|
|
8CX2
 
 | | Cryo-EM structure of human APOBEC3G/HIV-1 Vif/CBFbeta/ELOB/ELOC dimeric complex in State 2 | | Descriptor: | Core-binding factor subunit beta, DNA dC->dU-editing enzyme APOBEC-3G, Elongin-B, ... | | Authors: | Li, Y, Langley, C, Azumaya, C.M, Echeverria, I, Chesarino, N.M, Emerman, M, Cheng, Y, Gross, J.D. | | Deposit date: | 2022-05-19 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The structural basis for HIV-1 Vif antagonism of human APOBEC3G.
Nature, 615, 2023
|
|
8CX1
 
 | | Cryo-EM structure of human APOBEC3G/HIV-1 Vif/CBFbeta/ELOB/ELOC dimeric complex in State 1 | | Descriptor: | Core-binding factor subunit beta, DNA dC->dU-editing enzyme APOBEC-3G, Elongin-B, ... | | Authors: | Li, Y, Langley, C, Azumaya, C.M, Echeverria, I, Chesarino, N.M, Emerman, M, Cheng, Y, Gross, J.D. | | Deposit date: | 2022-05-19 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The structural basis for HIV-1 Vif antagonism of human APOBEC3G.
Nature, 615, 2023
|
|
4HUP
 
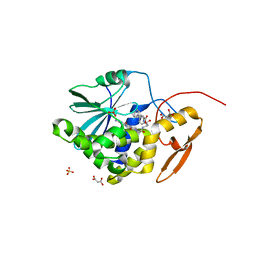 | | Structure of ricin A chain bound with N-(N-(pterin-7-yl)carbonylglycyl)-L-phenylalanyl)-L-phenylalanine | | Descriptor: | (2S)-2-[[(2S)-2-[2-[(2-azanyl-4-oxidanylidene-1H-pteridin-7-yl)carbonylamino]ethanoylamino]-3-phenyl-propanoyl]amino]-3-phenyl-propanoic acid, MALONIC ACID, Ricin, ... | | Authors: | Jasheway, K.R, Monzingo, A.F, Saito, R, Pruet, J.M, Manzano, L.A, Wiget, P.A, Anslyn, E.V, Robertus, J.D. | | Deposit date: | 2012-11-02 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Peptide-conjugated pterins as inhibitors of ricin toxin A.
J.Med.Chem., 56, 2013
|
|
7QC5
 
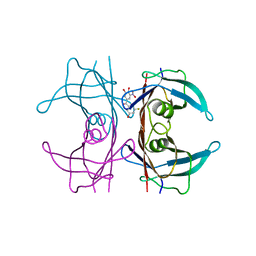 | | Crystal structure of human wild type transthyretin in complex with (3,4-dihydroxy-5-nitrophenyl)-(3-fluoro-5-hydroxyphenyl)methanone compound | | Descriptor: | (3-fluoranyl-5-oxidanyl-phenyl)-[3-nitro-4,5-bis(oxidanyl)phenyl]methanone, Transthyretin | | Authors: | Varejao, N, Pinheiro, F, Pallares, I, Ventura, S, Reverter, D. | | Deposit date: | 2021-11-22 | | Release date: | 2022-11-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Development of a Highly Potent Transthyretin Amyloidogenesis Inhibitor: Design, Synthesis, and Evaluation.
J.Med.Chem., 65, 2022
|
|
6YYE
 
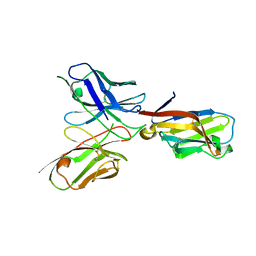 | | TREM2 extracellular domain (19-131) in complex with single-chain variable fragment (scFv-2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, TREM2 Single chain variable 2, Triggering receptor expressed on myeloid cells 2 | | Authors: | Szykowska, A, Preger, C, Krojer, T, Mukhopadhyay, S.M.M, McKinley, G, Graslund, S, Wigren, E, Persson, H, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Di Daniel, E, Burgess-Brown, N, Bullock, A. | | Deposit date: | 2020-05-04 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.36 Å) | | Cite: | Selection and structural characterization of anti-TREM2 scFvs that reduce levels of shed ectodomain.
Structure, 29, 2021
|
|
3OCH
 
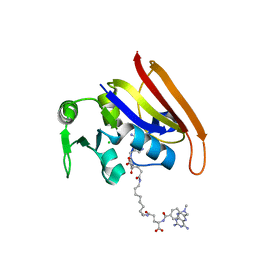 | | Chemically Self-assembled Antibody Nanorings (CSANs): Design and Characterization of an Anti-CD3 IgM Biomimetic | | Descriptor: | (2S,2'S)-5,5'-(nonane-1,9-diyldiimino)bis(2-{[(4-{[(2,4-diaminopteridin-6-yl)methyl](methyl)amino}phenyl)carbonyl]amino}-5-oxopentanoic acid) (non-preferred name), CHLORIDE ION, Dihydrofolate reductase, ... | | Authors: | Cody, V. | | Deposit date: | 2010-08-10 | | Release date: | 2010-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Chemically Self-Assembled Antibody Nanorings (CSANs): Design and Characterization of an Anti-CD3 IgM Biomimetic.
J.Am.Chem.Soc., 132, 2010
|
|
4D9R
 
 | |
8JCJ
 
 | |
8G59
 
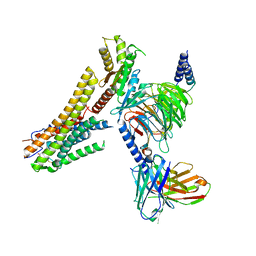 | | Cryo-EM structure of the TUG891 bound GPR120-Giq complex | | Descriptor: | 3-{4-[(4-fluoro-4'-methyl[1,1'-biphenyl]-2-yl)methoxy]phenyl}propanoic acid, Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Mao, C, Xiao, P, Tao, X, Qin, J, He, Q, Zhang, C, Yu, X, Zhang, Y, Sun, J. | | Deposit date: | 2023-02-12 | | Release date: | 2023-03-08 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Unsaturated bond recognition leads to biased signal in a fatty acid receptor.
Science, 380, 2023
|
|
8Q18
 
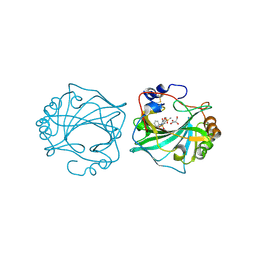 | | The Crystal Structure of Human Carbonic Anhydrase IX in Complex with Sulfonamide | | Descriptor: | 1,1,3-tris(oxidanylidene)-2-(2-phenylethyl)-1,2-benzothiazole-6-sulfonamide, Carbonic anhydrase 9, GLYCEROL, ... | | Authors: | Leitans, J, Tars, K. | | Deposit date: | 2023-07-31 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural Basis of Saccharin Derivative Inhibition of Carbonic Anhydrase IX.
Chemmedchem, 18, 2023
|
|
8Q19
 
 | | The Crystal Structure of Human Carbonic Anhydrase IX in Complex with Sulfonamide | | Descriptor: | 2-but-2-ynyl-1,1,3-tris(oxidanylidene)-1,2-benzothiazole-6-sulfonamide, Carbonic anhydrase 9, GLYCEROL, ... | | Authors: | Leitans, J, Tars, K. | | Deposit date: | 2023-07-31 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Structural Basis of Saccharin Derivative Inhibition of Carbonic Anhydrase IX.
Chemmedchem, 18, 2023
|
|
8Q1A
 
 | | The Crystal Structure of Human Carbonic Anhydrase IX in Complex with inhibitor | | Descriptor: | 1,1,3-tris(oxidanylidene)-2-pentyl-1,2-benzothiazole-6-sulfonamide, Carbonic anhydrase 9, GLYCEROL, ... | | Authors: | Leitans, J, Tars, K. | | Deposit date: | 2023-07-31 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis of Saccharin Derivative Inhibition of Carbonic Anhydrase IX.
Chemmedchem, 18, 2023
|
|
