4D6I
 
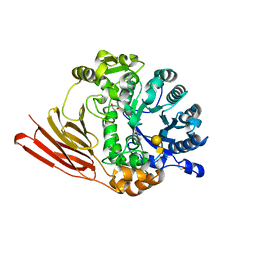 | | Crystal structure of a family 98 glycoside hydrolase catalytic module (Sp3GH98) in complex with the type 1 blood group A-tetrasaccharide (E558A L19 mutant) | | Descriptor: | 1,2-ETHANEDIOL, GLYCOSIDE HYDROLASE, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kwan, D.H, Constantinescu, I, Chapanian, R, Higgins, M.A, Samain, E, Boraston, A.B, Kizhakkedathu, J.N, Withers, S.G. | | Deposit date: | 2014-11-11 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Towards Efficient Enzymes for the Generation of Universal Blood Through Structure-Guided Directed Evolution.
J.Am.Chem.Soc., 137, 2015
|
|
8QD6
 
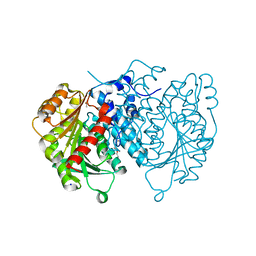 | | AntI Ser245DHA (PMSF) | | Descriptor: | ACETATE ION, Photorhabdus luminescens subsp. laumondii TTO1 complete genome segment 15/17, SODIUM ION, ... | | Authors: | Schmalhofer, M, Vagstad, A.L, Zhou, Q, Bode, H.B, Groll, M. | | Deposit date: | 2023-08-28 | | Release date: | 2024-03-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Polyketide Trimming Shapes Dihydroxynaphthalene-Melanin and Anthraquinone Pigments.
Adv Sci, 11, 2024
|
|
4D97
 
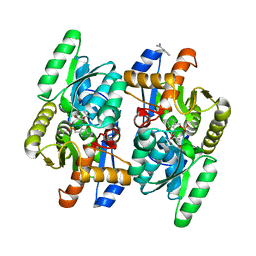 | | Salmonella typhimurium D-Cysteine desulfhydrase with D-ser bound at active site | | Descriptor: | BENZAMIDINE, D-SERINE, D-cysteine desulfhydrase | | Authors: | Bharath, S.R, Shveta, B, Rajesh, K.H, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2012-01-11 | | Release date: | 2012-05-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural and Mutational Studies on Substrate Specificity and Catalysis of Salmonella typhimurium D-Cysteine Desulfhydrase.
Plos One, 7, 2012
|
|
4D9W
 
 | |
4LI6
 
 | | TANKYRASE-1 Complexed with small molecule inhibitor N-[(4-oxo-3,4-dihydroquinazolin-2-yl)methyl]-3-phenyl-N-(thiophen-2-ylmethyl)propanamide | | Descriptor: | N-[(4-oxo-3,4-dihydroquinazolin-2-yl)methyl]-3-phenyl-N-(thiophen-2-ylmethyl)propanamide, SULFATE ION, Tankyrase-1, ... | | Authors: | Kirby, C.A, Stams, T. | | Deposit date: | 2013-07-02 | | Release date: | 2013-08-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Identification of NVP-TNKS656: The Use of Structure-Efficiency Relationships To Generate a Highly Potent, Selective, and Orally Active Tankyrase Inhibitor.
J.Med.Chem., 56, 2013
|
|
4D6H
 
 | | Crystal structure of a family 98 glycoside hydrolase catalytic module (Sp3GH98) in complex with the type 1 blood group A-tetrasaccharide (E558A X02 mutant) | | Descriptor: | 1,2-ETHANEDIOL, GLYCOSIDE HYDROLASE, SULFATE ION, ... | | Authors: | Kwan, D.H, Constantinescu, I, Chapanian, R, Higgins, M.A, Samain, E, Boraston, A.B, Kizhakkedathu, J.N, Withers, S.G. | | Deposit date: | 2014-11-11 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Towards Efficient Enzymes for the Generation of Universal Blood Through Structure-Guided Directed Evolution.
J.Am.Chem.Soc., 137, 2015
|
|
4LEY
 
 | | Structure of mouse cGAS bound to 18 bp DNA | | Descriptor: | 18 bp dsDNA, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Li, P. | | Deposit date: | 2013-06-26 | | Release date: | 2013-12-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Cyclic GMP-AMP Synthase Is Activated by Double-Stranded DNA-Induced Oligomerization.
Immunity, 39, 2013
|
|
8QTL
 
 | | Aplysia californica acetylcholine-binding protein in complex with Spiroimine (-)-4 S | | Descriptor: | CHLORIDE ION, ISOPROPYL ALCOHOL, Soluble acetylcholine receptor, ... | | Authors: | Sulzenbacher, G, Bourne, Y, Marchot, P. | | Deposit date: | 2023-10-12 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Cyclic Imine Core Common to the Marine Macrocyclic Toxins Is Sufficient to Dictate Nicotinic Acetylcholine Receptor Antagonism.
Mar Drugs, 22, 2024
|
|
4CRF
 
 | | Creating novel F1 inhibitors through fragment based lead generation and structure aided drug design | | Descriptor: | CHLORIDE ION, Coagulation factor XIa light chain, GLYCEROL, ... | | Authors: | Sandmark, J, Oster, L, Fjellstrom, O, Akkaya, S, Beisel, H.G, Eriksson, P.O, Erixon, K, Gustafsson, D, Jurva, U, Kang, D, Karis, D, Knecht, W, Nerme, V, Nilsson, I, Olsson, T, Redzic, A, Roth, R, Tigerstrom, A. | | Deposit date: | 2014-02-26 | | Release date: | 2015-02-11 | | Last modified: | 2022-05-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Creating Novel Activated Factor Xi Inhibitors Through Fragment Based Lead Generation and Structure Aided Drug Design.
Plos One, 10, 2015
|
|
8QX2
 
 | |
8QBH
 
 | | AntI in complex with 1-Naphthol | | Descriptor: | 1-NAPHTHOL, Photorhabdus luminescens subsp. laumondii TTO1 complete genome segment 15/17 | | Authors: | Schmalhofer, M, Vagstad, A.L, Zhou, Q, Bode, H.B, Groll, M. | | Deposit date: | 2023-08-24 | | Release date: | 2024-03-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Polyketide Trimming Shapes Dihydroxynaphthalene-Melanin and Anthraquinone Pigments.
Adv Sci, 11, 2024
|
|
4WA8
 
 | |
4WB0
 
 | |
4CRP
 
 | | Solution structure of a TrkAIg2 domain construct for use in drug discovery | | Descriptor: | HIGH AFFINITY NERVE GROWTH FACTOR RECEPTOR | | Authors: | Shoemark, D.K, Fahey, M, Williams, C, Sessions, R.B, Crump, M.P, Allen-Birt, S.J. | | Deposit date: | 2014-02-28 | | Release date: | 2015-01-14 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Design and Nuclear Magnetic Resonance (NMR) Structure Determination of the Second Extracellular Immunoglobulin Tyrosine Kinase a (Trkaig2) Domain Construct for Binding Site Elucidation in Drug Discovery
J.Med.Chem., 58, 2015
|
|
4LG7
 
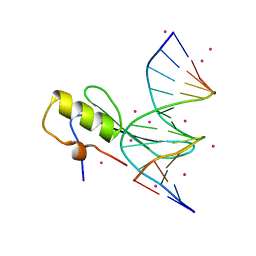 | | Crystal structure MBD4 MBD domain in complex with methylated CpG DNA | | Descriptor: | DNA (5'-D(*GP*CP*CP*AP*AP*(5CM)P*GP*TP*TP*GP*GP*C)-3'), Methyl-CpG-binding domain protein 4, UNKNOWN ATOM OR ION | | Authors: | Xu, C, Tempel, W, Wernimont, A.K, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-06-27 | | Release date: | 2013-07-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure MBD4 MBD domain in complex with methylated CpG DNA
To be Published
|
|
4D7Z
 
 | | E. coli L-aspartate-alpha-decarboxylase mutant N72Q to a resolution of 1.9 Angstroms | | Descriptor: | ASPARTATE 1-DECARBOXYLASE ALPHA CHAIN, ASPARTATE 1-DECARBOXYLASE BETA CHAIN, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bravo, J.P.K, Monteiro, D.C.F, Webb, M.E, Pearson, A.R. | | Deposit date: | 2014-12-02 | | Release date: | 2016-01-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Structure of the E. Coli L-Aspartate-Alpha-Decarboxylase Mutant N72Q to a Resolution of 1.9 Angstroms
To be Published
|
|
8QBI
 
 | | AntI in closed state | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Photorhabdus luminescens subsp. laumondii TTO1 complete genome segment 15/17, ... | | Authors: | Schmalhofer, M, Vagstad, A.L, Zhou, Q, Bode, H.B, Groll, M. | | Deposit date: | 2023-08-24 | | Release date: | 2024-03-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Polyketide Trimming Shapes Dihydroxynaphthalene-Melanin and Anthraquinone Pigments.
Adv Sci, 11, 2024
|
|
4UZJ
 
 | |
8QD5
 
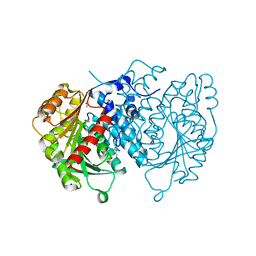 | | AntI Ser245DHA (PSF) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Photorhabdus luminescens subsp. laumondii TTO1 complete genome segment 15/17, ... | | Authors: | Schmalhofer, M, Vagstad, A.L, Zhou, Q, Bode, H.B, Groll, M. | | Deposit date: | 2023-08-28 | | Release date: | 2024-03-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Polyketide Trimming Shapes Dihydroxynaphthalene-Melanin and Anthraquinone Pigments.
Adv Sci, 11, 2024
|
|
4CMG
 
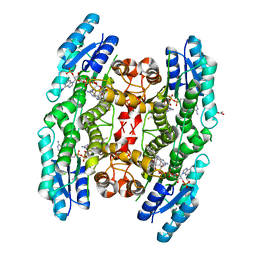 | | Crystal structure of pteridine reductase 1 (PTR1) from Trypanosoma brucei in ternary complex with cofactor and inhibitor | | Descriptor: | 6-(4-fluorophenyl)-5-(4-methoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidine-2,4-diamine, ACETATE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Barrack, K.L, Hunter, W.N. | | Deposit date: | 2014-01-16 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Design and Synthesis of Antiparasitic Pyrrolopyrimidines Targeting Pteridine Reductase 1.
J.Med.Chem., 57, 2014
|
|
4CO5
 
 | |
4CMC
 
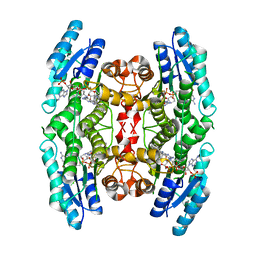 | | Crystal structure of pteridine reductase 1 (PTR1) from Trypanosoma brucei in ternary complex with cofactor and inhibitor | | Descriptor: | N4-cyclohexyl-5,6-diphenyl-7H-pyrrolo[2,3-d]pyrimidine-2,4-diamine, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PTERIDINE REDUCTASE 1 | | Authors: | Barrack, K.L, Hunter, W.N. | | Deposit date: | 2014-01-16 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Based Design and Synthesis of Antiparasitic Pyrrolopyrimidines Targeting Pteridine Reductase 1.
J.Med.Chem., 57, 2014
|
|
8QUD
 
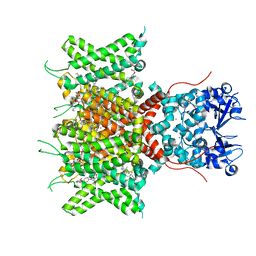 | | Cryo-EM Structure of Human Kv3.1 in Complex with Modulator AUT5 | | Descriptor: | (5R)-5-ethyl-3-(6-spiro[2H-1-benzofuran-3,1'-cyclopropane]-4-yloxypyridin-3-yl)imidazolidine-2,4-dione, 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Chi, G, Mckinley, G, Marsden, B, Pike, A.C.W, Ye, M, Brooke, L.M, Bakshi, S, Lakshminarayana, B, Pilati, N, Marasco, A, Gunthorpe, M, Alvaro, G.S, Large, C.H, Williams, E, Sauer, D.B. | | Deposit date: | 2023-10-16 | | Release date: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | The binding and mechanism of a positive allosteric modulator of Kv3 channels.
Nat Commun, 15, 2024
|
|
4LH1
 
 | |
4LH3
 
 | |
