2P9W
 
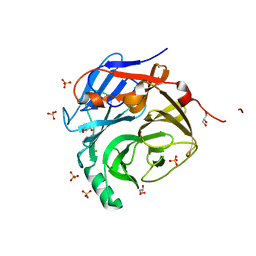 | | Crystal Structure of the Major Malassezia sympodialis Allergen Mala s 1 | | Descriptor: | CHLORIDE ION, GLYCEROL, Mal s 1 allergenic protein, ... | | Authors: | Vilhelmsson, M, Zargari, A, Crameri, R, Rasool, O, Achour, A, Scheynius, A, Hallberg, B.M. | | Deposit date: | 2007-03-26 | | Release date: | 2007-05-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal Structure of the Major Malassezia sympodialis Allergen Mala s 1 Reveals a beta-Propeller Fold: A Novel Fold Among Allergens.
J.Mol.Biol., 369, 2007
|
|
7MTP
 
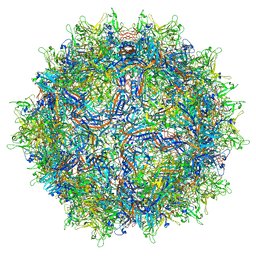 | | Structure of the adeno-associated virus 9 capsid at pH 5.5 | | Descriptor: | Capsid protein VP1 | | Authors: | Penzes, J.J, Chipman, P, Bhattacharya, N, Zeher, A, Huang, R, McKenna, R, Agbandje-McKenna, M. | | Deposit date: | 2021-05-13 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Adeno-associated Virus 9 Structural Rearrangements Induced by Endosomal Trafficking pH and Glycan Attachment.
J.Virol., 95, 2021
|
|
7MIK
 
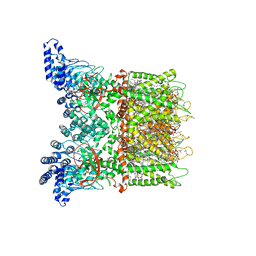 | | Mouse TRPV3 in MSP2N2 nanodiscs, closed state at 42 degrees Celsius | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, SODIUM ION, Transient receptor potential cation channel subfamily V member 3 | | Authors: | Nadezhdin, K.D, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2021-04-17 | | Release date: | 2021-07-21 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Structural mechanism of heat-induced opening of a temperature-sensitive TRP channel.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7MTW
 
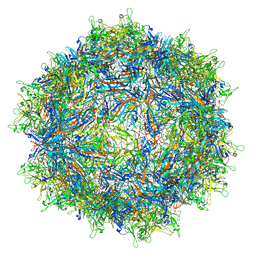 | | Structure of the adeno-associated virus 9 capsid at pH 4.0 | | Descriptor: | Capsid protein VP1 | | Authors: | Penzes, J.J, Chipman, P, Bhattacharya, N, Zeher, A, Huang, R, McKenna, R, Agbandje-McKenna, M. | | Deposit date: | 2021-05-13 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Adeno-associated Virus 9 Structural Rearrangements Induced by Endosomal Trafficking pH and Glycan Attachment.
J.Virol., 95, 2021
|
|
7MIN
 
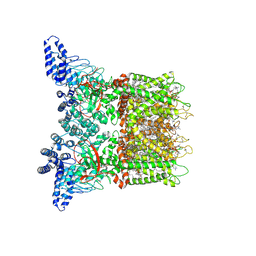 | | Mouse TRPV3 in cNW11 nanodiscs, closed state at 42 degrees Celsius | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, SODIUM ION, Transient receptor potential cation channel subfamily V member 3 | | Authors: | Nadezhdin, K.D, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2021-04-17 | | Release date: | 2021-07-21 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural mechanism of heat-induced opening of a temperature-sensitive TRP channel.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7DHF
 
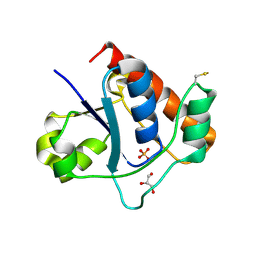 | | Vibrio vulnificus Wzb in complex with benzylphosphonate | | Descriptor: | GLYCEROL, PHOSPHATE ION, Protein-tyrosine-phosphatase | | Authors: | Ma, Q, Wang, X. | | Deposit date: | 2020-11-14 | | Release date: | 2021-01-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.211 Å) | | Cite: | Wzb of Vibrio vulnificus represents a new group of low-molecular-weight protein tyrosine phosphatases with a unique insertion in the W-loop.
J.Biol.Chem., 296, 2021
|
|
7MIM
 
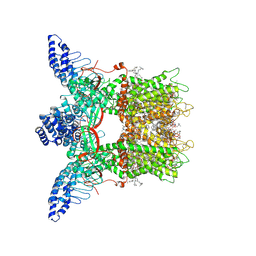 | | Mouse TRPV3 in cNW11 nanodiscs, closed state at 4 degrees Celsius | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, SODIUM ION, Transient receptor potential cation channel subfamily V member 3 | | Authors: | Neuberger, A, Nadezhdin, K.D, Sobolevsky, A.I. | | Deposit date: | 2021-04-17 | | Release date: | 2021-07-21 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Structural mechanism of heat-induced opening of a temperature-sensitive TRP channel.
Nat.Struct.Mol.Biol., 28, 2021
|
|
3H5V
 
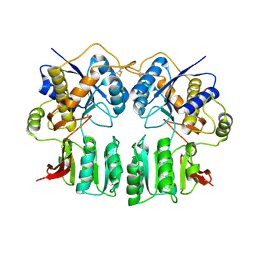 | | Crystal structure of the GluR2-ATD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 2 | | Authors: | Jin, R, Singh, S.K, Gu, S, Furukawa, H, Sobolevsky, A, Zhou, J, Jin, Y, Gouaux, E. | | Deposit date: | 2009-04-22 | | Release date: | 2009-06-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structure and association behaviour of the GluR2 amino-terminal domain.
Embo J., 28, 2009
|
|
2GYD
 
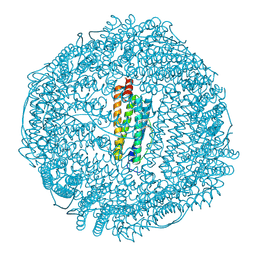 | | Complex of equine apoferritin with the H-diaziflurane photolabeling reagent | | Descriptor: | 2-(1,1-DIFLUOROETHOXY)-1,1,1-TRIFLUOROETHANE, CADMIUM ION, ferritin L subunit | | Authors: | Loll, P.J, Rossi, M.J, Eckenhoff, R. | | Deposit date: | 2006-05-09 | | Release date: | 2006-06-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Photoactive analogues of the haloether anesthetics provide high-resolution features from low-affinity interactions.
Acs Chem.Biol., 1, 2006
|
|
7CY4
 
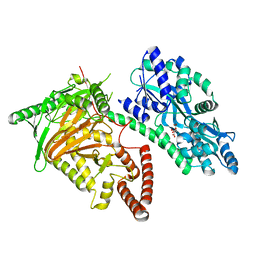 | | Crystal Structure of CMD1 in apo form | | Descriptor: | CITRIC ACID, FE (III) ION, Maltodextrin-binding protein,5-methylcytosine-modifying enzyme 1 | | Authors: | Li, W, Zhang, T, Sun, M, Ding, J. | | Deposit date: | 2020-09-03 | | Release date: | 2020-12-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular mechanism for vitamin C-derived C 5 -glyceryl-methylcytosine DNA modification catalyzed by algal TET homologue CMD1.
Nat Commun, 12, 2021
|
|
7MIL
 
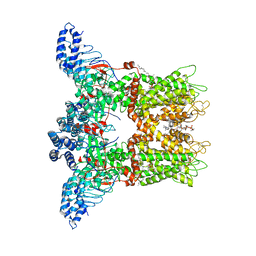 | | Mouse TRPV3 in MSP2N2 nanodiscs, sensitized state at 42 degrees Celsius | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, SODIUM ION, Transient receptor potential cation channel subfamily V member 3 | | Authors: | Nadezhdin, K.D, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2021-04-17 | | Release date: | 2021-07-21 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Structural mechanism of heat-induced opening of a temperature-sensitive TRP channel.
Nat.Struct.Mol.Biol., 28, 2021
|
|
3H7H
 
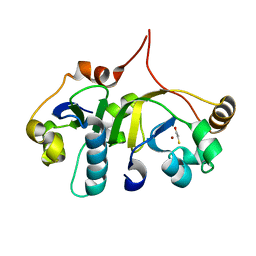 | | Crystal structure of the human transcription elongation factor DSIF, hSpt4/hSpt5 (176-273) | | Descriptor: | BETA-MERCAPTOETHANOL, Transcription elongation factor SPT4, Transcription elongation factor SPT5, ... | | Authors: | Wenzel, S, Wohrl, B.M, Rosch, P, Martins, B.M. | | Deposit date: | 2009-04-27 | | Release date: | 2009-12-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of the human transcription elongation factor DSIF hSpt4 subunit in complex with the hSpt5 dimerization interface
Biochem.J., 425, 2010
|
|
2P0I
 
 | | Crystal structure of L-rhamnonate dehydratase from Gibberella zeae | | Descriptor: | GLYCEROL, L-rhamnonate dehydratase, SULFATE ION | | Authors: | Patskovsky, Y, Toro, R, Sauder, J.M, Dickey, M, Logan, C, Gheyi, T, Wasserman, S.R, Smith, D, Gerlt, J, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-02-28 | | Release date: | 2007-03-13 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of L-Rhamnonate Dehydratase from Gibberella Zeae
To be Published
|
|
2HBW
 
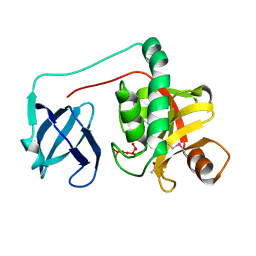 | |
7M3N
 
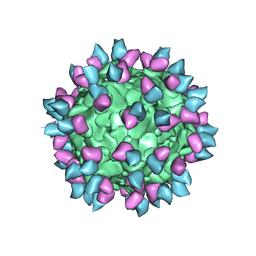 | |
3B8S
 
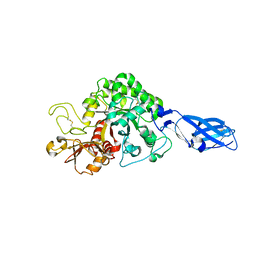 | | Crystal structure of wild-type chitinase A from Vibrio harveyi | | Descriptor: | Chitinase A | | Authors: | Songsiriritthigul, C, Pantoom, S, Aguda, A.H, Robinson, R.C, Suginta, W. | | Deposit date: | 2007-11-01 | | Release date: | 2008-04-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of Vibrio harveyi chitinase A complexed with chitooligosaccharides: implications for the catalytic mechanism
J.Struct.Biol., 162, 2008
|
|
2PDA
 
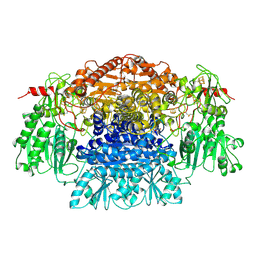 | |
2HCV
 
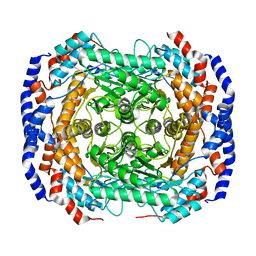 | | Crystal structure of L-rhamnose isomerase from Pseudomonas stutzeri with metal ion | | Descriptor: | L-rhamnose isomerase, ZINC ION | | Authors: | Yoshida, H, Yamada, M, Takada, G, Izumori, K, Kamitori, S. | | Deposit date: | 2006-06-19 | | Release date: | 2006-12-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structures of l-Rhamnose Isomerase from Pseudomonas stutzeri in Complexes with l-Rhamnose and d-Allose Provide Insights into Broad Substrate Specificity
J.Mol.Biol., 365, 2007
|
|
3GN0
 
 | |
7D3F
 
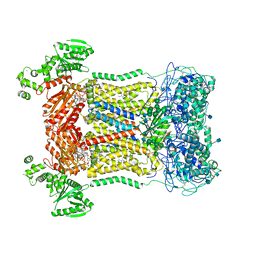 | | Cryo-EM structure of human DUOX1-DUOXA1 in high-calcium state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Dual oxidase 1, ... | | Authors: | Chen, L, Wu, J.X. | | Deposit date: | 2020-09-19 | | Release date: | 2020-12-09 | | Last modified: | 2021-06-23 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structures of human dual oxidase 1 complex in low-calcium and high-calcium states.
Nat Commun, 12, 2021
|
|
7D7Z
 
 | |
7MKF
 
 | |
7DC7
 
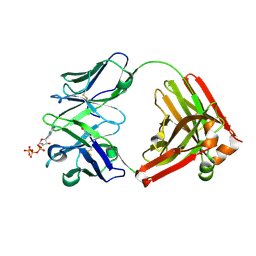 | | Crystal structure of D12 Fab-ATP complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, D12 Fab heavy chain, D12 Fab light chain | | Authors: | Kawauchi, H, Fukami, T.A, Tatsumi, K, Torizawa, T, Mimoto, F. | | Deposit date: | 2020-10-23 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Exploitation of Elevated Extracellular ATP to Specifically Direct Antibody to Tumor Microenvironment.
Cell Rep, 33, 2020
|
|
7M3L
 
 | |
3HBH
 
 | |
