2BAA
 
 | | THE REFINED CRYSTAL STRUCTURE OF AN ENDOCHITINASE FROM HORDEUM VULGARE L. SEEDS TO 1.8 ANGSTROMS RESOLUTION | | Descriptor: | ENDOCHITINASE (26 KD) | | Authors: | Hart, P.J, Pfluger, H.D, Monzingo, A.F, Ready, M.P, Ernst, S.R, Hollis, T, Robertus, J.D. | | Deposit date: | 1995-01-26 | | Release date: | 1996-01-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The refined crystal structure of an endochitinase from Hordeum vulgare L. seeds at 1.8 A resolution.
J.Mol.Biol., 248, 1995
|
|
3SBW
 
 | | Crystal structure of the complex between the extracellular domains of mouse PD-1 mutant and human PD-L1 | | Descriptor: | Programmed cell death 1 ligand 1, Programmed cell death protein 1 | | Authors: | Lazar-Molnar, E, Ramagopal, U.A, Cao, E, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2011-06-06 | | Release date: | 2011-07-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of the complex between the extracellular domains of mouse PD-1 mutant and human PD-L1
To be published
|
|
7F82
 
 | | Structure of the bacterial cellulose synthase subunit Z in complex with cellooligosaccharides from Enterobacter sp. CJF-002 | | Descriptor: | Glucanase, S,R MESO-TARTARIC ACID, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Fujiwara, T, Fujishima, A, Yao, M. | | Deposit date: | 2021-06-30 | | Release date: | 2022-02-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural snapshot of a glycoside hydrolase family 8 endo-beta-1,4-glucanase capturing the state after cleavage of the scissile bond.
Acta Crystallogr.,Sect.D, 78, 2022
|
|
1O8Y
 
 | | Solution structure of SFTI-1(6,5), an acyclic permutant of the proteinase inhibitor SFTI-1, trans-trans-trans conformer (tt-A) | | Descriptor: | CYCLIC TRYPSIN INHIBITOR | | Authors: | Marx, U.C, Craik, D.J. | | Deposit date: | 2002-12-09 | | Release date: | 2003-03-13 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Enzymatic Cyclization of a Potent Bowman-Birk Protease Inhibitor, Sunflower Trypsin Inhibitor-1, and Solution Structure of an Acyclic Precursor Peptide
J.Biol.Chem., 278, 2003
|
|
2XBV
 
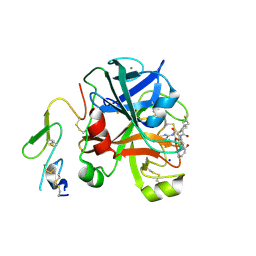 | | Factor Xa in complex with a pyrrolidine-3,4-dicarboxylic acid inhibitor | | Descriptor: | (3R,4R)-1-(2,2-DIFLUORO-ETHYL)-PYRROLIDINE-3,4-DICARBOXYLIC ACID 3-[(5-CHLORO-PYRIDIN-2-YL)-AMIDE]-4-{[2-FLUORO-4-(2-OXO-2H-PYRIDIN-1-YL)-PHENYL]-AMIDE}, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION, ... | | Authors: | Banner, D.W, Benz, J, Schlatter, D, Anselm, L, Haap, W. | | Deposit date: | 2010-04-15 | | Release date: | 2010-08-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Discovery of a Factor Xa Inhibitor (3R,4R)-1-(2,2-Difluoro-Ethyl)-Pyrrolidine-3,4-Dicarboxylic Acid 3-[(5-Chloro-Pyridin-2-Yl)-Amide] 4-{[2-Fluoro-4-(2-Oxo-2H-Pyridin-1-Yl)-Phenyl]-Amide} as a Clinical Candidate.
Bioorg.Med.Chem., 20, 2010
|
|
1OJK
 
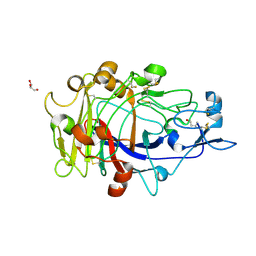 | | Anatomy of glycosynthesis: Structure and kinetics of the Humicola insolens Cel7BE197A and E197S glycosynthase mutants | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDOGLUCANASE I, GLYCEROL, ... | | Authors: | Ducros, V.M.-A, Tarling, C.A, Zechel, D.L, Brzozowski, A.M, Frandsen, T.P, Von Ossowski, I, Schulein, M, Withers, S.G, Davies, G.J. | | Deposit date: | 2003-07-10 | | Release date: | 2004-01-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Anatomy of Glycosynthesis: Structure and Kinetics of the Humicola Insolens Cel7B E197A and E197S Glycosynthase Mutants
Chem.Biol., 10, 2003
|
|
4FAM
 
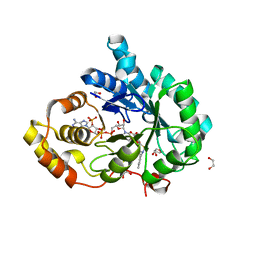 | | Crystal structure of human 17beta-hydroxysteroid dehydrogenase type 5 in complex with 3-((3,4-dihydroisoquinolin-2(1H)-yl)sulfonyl)benzoic acid (17) | | Descriptor: | 1,2-ETHANEDIOL, 3-(3,4-dihydroisoquinolin-2(1H)-ylsulfonyl)benzoic acid, Aldo-keto reductase family 1 member C3, ... | | Authors: | Turnbull, A.P, Jamieson, S.M.F, Brooke, D.G, Heinrich, D, Atwell, G.J, Silva, S, Hamilton, E.J, Rigoreau, L.J.M, Trivier, E, Soudy, C, Samlal, S.S, Owen, P.J, Schroeder, E, Raynham, T, Flanagan, J.U, Denny, W.A. | | Deposit date: | 2012-05-22 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 3-(3,4-Dihydroisoquinolin-2(1H)-ylsulfonyl)benzoic acids; a New Class of Highly Potent and Selective Inhibitors of the Type 5 17-beta-hydroxysteroid Dehydrogenase AKR1C3
J.Med.Chem., 55, 2012
|
|
3UJ0
 
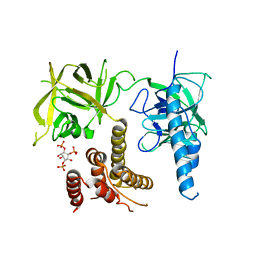 | | Crystal structure of the inositol 1,4,5-trisphosphate receptor with ligand bound form. | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol 1,4,5-trisphosphate receptor type 1 | | Authors: | Ikura, M, Seo, M.D, Ishiyama, N, Stathopulos, P. | | Deposit date: | 2011-11-07 | | Release date: | 2012-02-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural and functional conservation of key domains in InsP3 and ryanodine receptors.
Nature, 483, 2012
|
|
2AAY
 
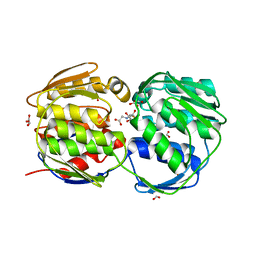 | | EPSP synthase liganded with shikimate and glyphosate | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, 3-phosphoshikimate 1-carboxyvinyltransferase, FORMIC ACID, ... | | Authors: | Priestman, M.A, Healy, M.L, Funke, T, Becker, A. | | Deposit date: | 2005-07-14 | | Release date: | 2006-02-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Molecular basis for the glyphosate-insensitivity of the reaction of 5-enolpyruvylshikimate 3-phosphate synthase with shikimate.
Febs Lett., 579, 2005
|
|
4G2H
 
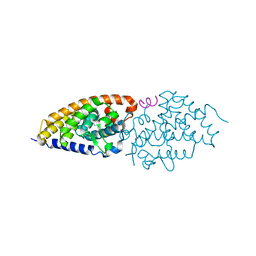 | | Structural basis for the accommodation of bis- and tris-aromatic derivatives in Vitamin D Nuclear Receptor | | Descriptor: | (3E,5E)-6-(3-{2-[3,4-bis(hydroxymethyl)phenyl]ethyl}phenyl)-1,1,1-trifluoro-2-(trifluoromethyl)octa-3,5-dien-2-ol, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | Authors: | Ciesielski, F, Sato, Y, Moras, D, Rochel, N. | | Deposit date: | 2012-07-12 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the accommodation of bis- and tris-aromatic derivatives in vitamin d nuclear receptor.
J.Med.Chem., 55, 2012
|
|
1D3E
 
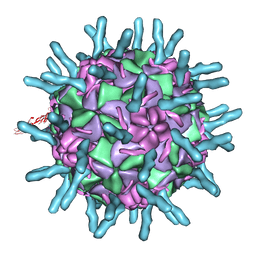 | |
1G97
 
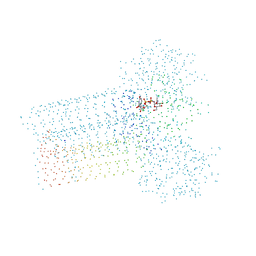 | | S.PNEUMONIAE GLMU COMPLEXED WITH UDP-N-ACETYLGLUCOSAMINE AND MG2+ | | Descriptor: | MAGNESIUM ION, N-ACETYLGLUCOSAMINE-1-PHOSPHATE URIDYLTRANSFERASE, SODIUM ION, ... | | Authors: | Kostrewa, D, D'Arcy, A, Kamber, M. | | Deposit date: | 2000-11-22 | | Release date: | 2001-05-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structures of Streptococcus pneumoniae N-acetylglucosamine-1-phosphate uridyltransferase, GlmU, in apo form at 2.33 A resolution and in complex with UDP-N-acetylglucosamine and Mg(2+) at 1.96 A resolution.
J.Mol.Biol., 305, 2001
|
|
2B4E
 
 | |
2X7U
 
 | | Structures of human carbonic anhydrase II inhibitor complexes reveal a second binding site for steroidal and non-steroidal inhibitors. | | Descriptor: | (9BETA,14BETA,17BETA)-17-HYDROXY-2-METHOXYESTRA-1,3,5(10)-TRIEN-3-YL SULFAMATE, CARBONIC ANHYDRASE 2, GLYCEROL, ... | | Authors: | Cozier, G.E, Leese, M.P, Lloyd, M.D, Baker, M.D, Thiyagarajan, N, Acharya, K.R, Potter, B.V.L. | | Deposit date: | 2010-03-03 | | Release date: | 2010-03-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structures of Human Carbonic Anhydrase II/Inhibitor Complexes Reveal a Second Binding Site for Steroidal and Non-Steroidal Inhibitors.
Biochemistry, 49, 2010
|
|
2N1A
 
 | | Docked structure between SUMO1 and ZZ-domain from CBP | | Descriptor: | CREB-binding protein, Small ubiquitin-related modifier 1, ZINC ION | | Authors: | Diehl, C. | | Deposit date: | 2015-03-26 | | Release date: | 2016-05-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Analysis of a Complex between Small Ubiquitin-like Modifier 1 (SUMO1) and the ZZ Domain of CREB-binding Protein (CBP/p300) Reveals a New Interaction Surface on SUMO.
J.Biol.Chem., 291, 2016
|
|
4CPZ
 
 | | Structure of the Neuraminidase from the B/Lyon/CHU/15.216/2011 virus in complex with Zanamivir | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Vachieri, S.G, Collins, P.J, Escuret, V, Casalegno, J.S, Cattle, N, Ferraris, O, Sabatier, M, Frobert, E, Caro, V, Skehel, J.J, Gamblin, S.J, Valla, F, Valette, M, Ottmann, M, McCauley, J.W, Daniels, R.S, Lina, B. | | Deposit date: | 2014-02-09 | | Release date: | 2014-05-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Novel I221 L Substitution in Neuraminidase Confers High Level Resistance to Oseltamivir in Influenza B Viruses.
J.Infect.Dis., 210, 2014
|
|
1Z84
 
 | | X-ray structure of galt-like protein from arabidopsis thaliana at5g18200 | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, ZINC ION, ... | | Authors: | Mccoy, J.G, Bitto, E, Phillips Jr, G.N, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-03-29 | | Release date: | 2005-04-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure and Mechanism of an ADP-Glucose Phosphorylase from
Arabidopsis thaliana
Biochemistry, 45, 2006
|
|
2ASF
 
 | | Crystal structure of the conserved hypothetical protein Rv2074 from Mycobacterium tuberculosis 1.6 A | | Descriptor: | CITRIC ACID, Hypothetical protein Rv2074, SODIUM ION | | Authors: | Biswal, B.K, Au, K, Cherney, M.M, Garen, C, James, M.N, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2005-08-23 | | Release date: | 2005-10-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The molecular structure of Rv2074, a probable pyridoxine 5'-phosphate oxidase from Mycobacterium tuberculosis, at 1.6 angstroms resolution.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
2AZA
 
 | |
2MU2
 
 | | NMR structure of the cap domain of NP_346487.1, a putative phosphoglycolate phosphatase from Streptococcus pneumoniae TIGR4 | | Descriptor: | Hydrolase, haloacid dehalogenase-like family | | Authors: | Jaudzems, K, Serrano, P, Pedrini, B, Geralt, M, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2014-09-03 | | Release date: | 2014-09-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | J-UNIO protocol used for NMR structure determination of the 206-residue protein NP_346487.1 from Streptococcus pneumoniae TIGR4.
J.Biomol.Nmr, 61, 2015
|
|
1ZGB
 
 | | Crystal Structure of Torpedo Californica Acetylcholinesterase in Complex With an (R)-Tacrine(10)-Hupyridone Inhibitor. | | Descriptor: | (5R)-5-{[10-(1,2,3,4-TETRAHYDROACRIDIN-9-YLAMINO)DECYL]AMINO}-5,6,7,8-TETRAHYDROQUINOLIN-2(1H)-ONE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase | | Authors: | Haviv, H, Wong, D.M, Greenblatt, H.M, Carlier, P.R, Pang, Y.P, Silman, I, Sussman, J.L, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2005-04-21 | | Release date: | 2005-08-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Packing Mediates Enantioselective Ligand Recognition at the Peripheral Site of Acetylcholinesterase
J.Am.Chem.Soc., 127, 2005
|
|
1QBB
 
 | | BACTERIAL CHITOBIASE COMPLEXED WITH CHITOBIOSE (DINAG) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHITOBIASE, SULFATE ION | | Authors: | Tews, I, Perrakis, A, Oppenheim, A, Dauter, Z, Wilson, K.S, Vorgias, C.E. | | Deposit date: | 1996-06-07 | | Release date: | 1997-02-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bacterial chitobiase structure provides insight into catalytic mechanism and the basis of Tay-Sachs disease.
Nat.Struct.Biol., 3, 1996
|
|
3ZV7
 
 | | Torpedo californica Acetylcholinesterase Inhibition by Bisnorcymserine | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bartolucci, C, Stojan, J, Greig, N.H, Lamba, D. | | Deposit date: | 2011-07-24 | | Release date: | 2012-05-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Kinetics of Torpedo Californica Acetylcholinesterase Inhibition by Bisnorcymserine and Crystal Structure of the Complex with its Leaving Group.
Biochem.J., 444, 2012
|
|
3SHJ
 
 | | Proteasome in complex with hydroxyurea derivative HU10 | | Descriptor: | 1-hydroxy-1-[(2R)-4-{3-[(3S,5S,7S)-tricyclo[3.3.1.1~3,7~]dec-1-yloxy]phenyl}but-3-yn-2-yl]urea, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Proteasome component C1, ... | | Authors: | Gallastegui, N, Beck, P, Arciniega, M, Hillebrand, S, Huber, R, Groll, M. | | Deposit date: | 2011-06-16 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Hydroxyureas as noncovalent proteasome inhibitors.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
3SA3
 
 | | Crystal structure of wild-type HIV-1 protease in complex with AG23 | | Descriptor: | ACETATE ION, N~2~-acetyl-N-[(2S,3R)-4-{(1,3-benzothiazol-6-ylsulfonyl)[(2S)-2-methylbutyl]amino}-3-hydroxy-1-phenylbutan-2-yl]-L-isoleucinamide, PHOSPHATE ION, ... | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2011-06-02 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Protease Inhibitors that protrude out from substrate envelope are more susceptible to developing drug resistance
To be Published
|
|
