1H5T
 
 | | Thymidylyltransferase complexed with Thymidylyldiphosphate-glucose | | Descriptor: | 2'DEOXY-THYMIDINE-5'-DIPHOSPHO-ALPHA-D-GLUCOSE, Glucose-1-phosphate thymidylyltransferase 1, SULFATE ION, ... | | Authors: | Rosano, C, Zuccotti, S, Bolognesi, M. | | Deposit date: | 2001-05-25 | | Release date: | 2001-11-23 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Kinetic and Crystallographic Analyses Support a Sequential-Ordered Bi Bi Catalytic Mechanism for Escherichia Coli Glucose-1-Phosphate Thymidylyltransferase
J.Mol.Biol., 313, 2001
|
|
4HRD
 
 | | Crystal structure of yeast 20S proteasome in complex with the natural product carmaphycin A | | Descriptor: | N-[(2S)-1-({(2S)-1-{[(2R,3S,4S)-1,3-dihydroxy-2,6-dimethylheptan-4-yl]amino}-4-[(R)-methylsulfinyl]-1-oxobutan-2-yl}amino)-3-methyl-1-oxobutan-2-yl]hexanamide, Proteasome component C1, Proteasome component C11, ... | | Authors: | Trivella, D.B.B, Stein, M, Groll, M. | | Deposit date: | 2012-10-27 | | Release date: | 2014-01-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Enzyme inhibition by hydroamination: design and mechanism of a hybrid carmaphycin-syringolin enone proteasome inhibitor.
Chem.Biol., 21, 2014
|
|
1HF6
 
 | | ENDOGLUCANASE CEL5A FROM BACILLUS AGARADHAERENS IN THE ORTHORHOMBIC CRYSTAL FORM IN COMPLEX WITH CELLOTRIOSE | | Descriptor: | ACETIC ACID, ENDOGLUCANASE B, GLYCEROL, ... | | Authors: | Varrot, A, Withers, S, Vasella, A, Schulein, M, Davies, G.J. | | Deposit date: | 2000-11-29 | | Release date: | 2001-11-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Direct Experimental Observation of the Hydrogen-Bonding Network of a Glycosidase Along its Reaction Coordinate Revealed by Atomic Resolution Analyses of Endoglucanase Cel5A
Acta Crystallogr.,Sect.D, 59, 2003
|
|
2C78
 
 | | EF-Tu complexed with a GTP analog and the antibiotic pulvomycin | | Descriptor: | (1S,2S,3E,5E,7E,10S,11S,12S)-12-[(2R,4E,6E,8Z,10R,12E,14E,16Z,18S,19Z)-10,18-DIHYDROXY-12,16,19-TRIMETHYL-11,22-DIOXOOX ACYCLODOCOSA-4,6,8,12,14,16,19-HEPTAEN-2-YL]-2,11-DIHYDROXY-1,10-DIMETHYL-9-OXOTRIDECA-3,5,7-TRIEN-1-YL 6-DEOXY-2,4-DI-O-METHYL-BETA-L-GALACTOPYRANOSIDE, ELONGATION FACTOR TU-A, MAGNESIUM ION, ... | | Authors: | Parmeggiani, A, Krab, I.M, Okamura, S, Nielsen, R.C, Nyborg, J, Nissen, P. | | Deposit date: | 2005-11-18 | | Release date: | 2006-03-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis of the action of pulvomycin and GE2270 A on elongation factor Tu.
Biochemistry, 45, 2006
|
|
4MZE
 
 | | Crystal structure of hPIV3 hemagglutinin-neuraminidase, H552Q/Q559R mutant | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Xu, R, Wilson, I.A. | | Deposit date: | 2013-09-30 | | Release date: | 2013-11-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Interaction between the hemagglutinin-neuraminidase and fusion glycoproteins of human parainfluenza virus type III regulates viral growth in vivo.
MBio, 4, 2013
|
|
2L7W
 
 | |
3GBN
 
 | | Crystal Structure of Fab CR6261 in Complex with the 1918 H1N1 influenza virus hemagglutinin | | Descriptor: | 1,2-ETHANEDIOL, 2-ETHOXYETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ekiert, D.C, Elsliger, M.A, Wilson, I.A. | | Deposit date: | 2009-02-20 | | Release date: | 2009-03-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Antibody recognition of a highly conserved influenza virus epitope.
Science, 324, 2009
|
|
2WT0
 
 | | Galectin domain of porcine adenovirus type 4 NADC-1 isolate fibre complexed with N-acetyl-lactosamine | | Descriptor: | NITRATE ION, PUTATIVE FIBER PROTEIN, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Guardado-Calvo, P, Munoz, E.M, Llamas-Saiz, A.L, Fox, G.C, Glasgow, J.N, van Raaij, M.J. | | Deposit date: | 2009-09-10 | | Release date: | 2010-08-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystallographic structure of porcine adenovirus type 4 fiber head and galectin domains.
J. Virol., 84, 2010
|
|
2WSV
 
 | | Galectin domain of porcine adenovirus type 4 NADC-1 isolate fibre complexed with lactose | | Descriptor: | NITRATE ION, PUTATIVE FIBER PROTEIN, beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Guardado-Calvo, P, Munoz, E.M, Llamas-Saiz, A.L, Fox, G.C, Glasgow, J.N, van Raaij, M.J. | | Deposit date: | 2009-09-10 | | Release date: | 2010-08-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic structure of porcine adenovirus type 4 fiber head and galectin domains.
J. Virol., 84, 2010
|
|
3WQ6
 
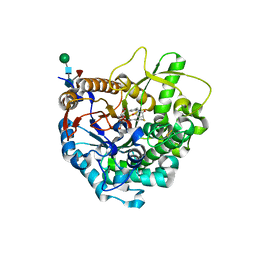 | |
2BZG
 
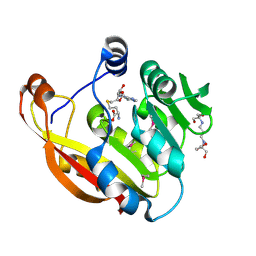 | | Crystal structure of thiopurine S-methyltransferase. | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, S-ADENOSYL-L-HOMOCYSTEINE, THIOPURINE S-METHYLTRANSFERASE | | Authors: | Battaile, K.P, Wu, H, Zeng, H, Loppnau, P, Dong, A, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-08-17 | | Release date: | 2005-08-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural Basis of Allele Variation of Human Thiopurine-S-Methyltransferase.
Proteins, 67, 2007
|
|
2BHU
 
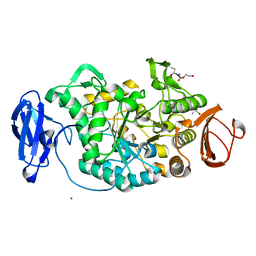 | | Crystal structure of Deinococcus radiodurans maltooligosyltrehalose trehalohydrolase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, MALTOOLIGOSYLTREHALOSE TREHALOHYDROLASE, ... | | Authors: | Timmins, J, Leiros, H.-K.S, Leonard, G, Leiros, I, McSweeney, S. | | Deposit date: | 2005-01-18 | | Release date: | 2005-03-31 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal Structure of Maltooligosyltrehalose Trehalohydrolase from Deinococcus Radiodurans in Complex with Disaccharides
J.Mol.Biol., 347, 2005
|
|
3OTU
 
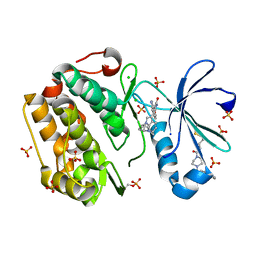 | | PDK1 mutant bound to allosteric disulfide fragment activator JS30 | | Descriptor: | 3-(1H-INDOL-3-YL)-4-{1-[2-(1-METHYLPYRROLIDIN-2-YL)ETHYL]-1H-INDOL-3-YL}-1H-PYRROLE-2,5-DIONE, 3-phosphoinositide-dependent protein kinase 1, 4-[4-(naphthalen-1-ylmethyl)piperazin-1-yl]-4-oxobutane-1-thiol, ... | | Authors: | Sadowsky, J.D, Wells, J.A. | | Deposit date: | 2010-09-13 | | Release date: | 2011-03-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1013 Å) | | Cite: | Turning a protein kinase on or off from a single allosteric site via disulfide trapping.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1OC5
 
 | | D405N mutant of the CELLOBIOHYDROLASE CEL6A FROM HUMICOLA INSOLENS in complex with methyl-cellobiosyl-4-deoxy-4-thio-beta-D-cellobioside | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CELLOBIOHYDROLASE II, GLYCEROL, ... | | Authors: | Varrot, A, Frandsen, T.P, Von Ossowski, I, Boyer, V, Driguez, H, Schulein, M, Davies, G.J. | | Deposit date: | 2003-02-06 | | Release date: | 2003-07-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for Ligand Binding and Processivity in Cellobiohydrolase Cel6A from Humicola Insolens
Structure, 11, 2003
|
|
3I3D
 
 | | E. COLI (lacZ) BETA-GALACTOSIDASE (M542A) IN COMPLEX WITH IPTG | | Descriptor: | 1-methylethyl 1-thio-beta-D-galactopyranoside, Beta-galactosidase, DIMETHYL SULFOXIDE, ... | | Authors: | Dugdale, M.L, Dymianiw, D, Minhas, B, Huber, R.E. | | Deposit date: | 2009-06-30 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Role of Met-542 as a guide for the conformational changes of Phe-601 that occur during the reaction of β-galactosidase (Escherichia coli).
Biochem.Cell Biol., 88, 2010
|
|
5QGF
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMOOA000539a | | Descriptor: | 1-[(3S,3aS,8bS)-7-chloro-3-(hydroxymethyl)-2,3,3a,8b-tetrahydro-1H-[1]benzofuro[3,2-b]pyrrol-1-yl]ethan-1-one, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.513 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
4GPT
 
 | | Crystal structure of KPT251 in complex with CRM1-Ran-RanBP1 | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-{3-[3,5-bis(trifluoromethyl)phenyl]-1H-1,2,4-triazol-1-yl}ethyl)-1,3,4-oxadiazole, CHLORIDE ION, ... | | Authors: | Sun, Q, Chook, Y.M. | | Deposit date: | 2012-08-21 | | Release date: | 2012-09-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Antileukemic activity of nuclear export inhibitors that spare normal hematopoietic cells.
Leukemia, 27, 2013
|
|
3ZMR
 
 | | Bacteroides ovatus GH5 xyloglucanase in complex with a XXXG heptasaccharide | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CELLULASE (GLYCOSYL HYDROLASE FAMILY 5), ... | | Authors: | Larsbrink, J, Rogers, T.E, Hemsworth, G.R, McKee, L.S, Spadiut, O, Klinter, S, Pudlo, N.A, Urs, K, Kelly, A.G, Cederholm, S.N, Davies, G.J, Martens, E.C, Brumer, H. | | Deposit date: | 2013-02-12 | | Release date: | 2014-01-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | A Discrete Genetic Locus Confers Xyloglucan Metabolism in Select Human Gut Bacteroidetes
Nature, 506, 2014
|
|
1T8U
 
 | | Crystal Structure of human 3-O-Sulfotransferase-3 with bound PAP and tetrasaccharide substrate | | Descriptor: | 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, ADENOSINE-3'-5'-DIPHOSPHATE, SODIUM ION, ... | | Authors: | Moon, A.F, Edavettal, S.C, Krahn, J.M, Munoz, E.M, Negishi, M, Linhardt, R.J, Liu, J, Pedersen, L.C. | | Deposit date: | 2004-05-13 | | Release date: | 2004-08-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural analysis of the sulfotransferase (3-o-sulfotransferase isoform 3) involved in the biosynthesis of an entry receptor for herpes simplex virus 1
J.Biol.Chem., 279, 2004
|
|
2BPM
 
 | | STRUCTURE OF CDK2-CYCLIN A WITH PHA-630529 | | Descriptor: | (2S)-N-[(3Z)-5-CYCLOPROPYL-3H-PYRAZOL-3-YLIDENE]-2-[4-(2-OXOIMIDAZOLIDIN-1-YL)PHENYL]PROPANAMIDE, CELL DIVISION PROTEIN KINASE 2, CYCLIN A2, ... | | Authors: | Cameron, A, Fogliatto, G, Pevarello, P, Brasca, M.G, Orsini, P, Traquandi, G, Longo, A, Nesi, M, Orzi, F, Piutti, C, Sansonna, P, Varasi, M, Vulpetti, A, Roletto, F, Alzani, R, Ciomei, M, Albanese, C, Pastori, W, Marsiglio, A, Pesenti, E, Fiorentini, F, Bischoff, J.R, Mercurio, C. | | Deposit date: | 2005-04-21 | | Release date: | 2005-12-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 3-Aminopyrazole Inhibitors of Cdk2-Cyclin a as Antitumor Agents. 2. Lead Optimization
J.Med.Chem., 48, 2005
|
|
1NFV
 
 | | X-ray structure of Desulfovibrio desulfuricans bacterioferritin: the diiron centre in different catalytic states (as-isolated structure) | | Descriptor: | 1,3,5,8-TETRAMETHYL-PORPHINE-2,4,6,7-TETRAPROPIONIC ACID FERROUS COMPLEX, 3-HYDROXYPYRUVIC ACID, FE (III) ION, ... | | Authors: | Macedo, S, Romao, C.V, Mitchell, E, Matias, P.M, Liu, M.Y, Xavier, A.V, LeGall, J, Teixeira, M, Lindley, P, Carrondo, M.A. | | Deposit date: | 2002-12-16 | | Release date: | 2003-04-01 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The nature of the di-iron site in the bacterioferritin from
Desulfovibrio desulfuricans
NAT.STRUCT.BIOL., 10, 2003
|
|
2Y5E
 
 | | BARLEY LIMIT DEXTRINASE IN COMPLEX WITH ALPHA-CYCLODEXTRIN | | Descriptor: | CALCIUM ION, Cyclohexakis-(1-4)-(alpha-D-glucopyranose), GLYCEROL, ... | | Authors: | Vester-Christensen, M.B, Hachem, M.A, Svensson, B, Henriksen, A. | | Deposit date: | 2011-01-13 | | Release date: | 2011-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Crystal Structure of an Essential Enzyme in Seed Starch Degradation: Barley Limit Dextrinase in Complex with Cyclodextrins.
J.Mol.Biol., 403, 2010
|
|
1XCW
 
 | | Acarbose Rearrangement Mechanism Implied by the Kinetic and Structural Analysis of Human Pancreatic alpha-Amylase in Complex with Analogues and Their Elongated Counterparts | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Alpha-amylase, ... | | Authors: | Li, C, Begum, A, Numao, S, Park, K.H, Withers, S.G, Brayer, G.D. | | Deposit date: | 2004-09-03 | | Release date: | 2004-12-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Acarbose Rearrangement Mechanism Implied by the Kinetic and Structural Analysis of Human Pancreatic alpha-Amylase in Complex with Analogues and Their Elongated Counterparts
Biochemistry, 44, 2005
|
|
2LV2
 
 | | Solution NMR structure of C2H2-type Zinc-fingers 4 and 5 from human Insulinoma-associated protein 1 (fragment 424-497), Northeast Structural Genomics Consortium Target HR7614B | | Descriptor: | Insulinoma-associated protein 1, ZINC ION | | Authors: | Yang, Y, Ramelot, T.A, Cort, J.R, Shastry, R, Kohan, E, Janjua, H, Xiao, R, Acton, T, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-06-27 | | Release date: | 2012-10-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of C2H2-type Zinc-fingers 4 and 5 from human Insulinoma-associated protein 1 (fragment 424-497), Northeast Structural
Genomics Consortium Target HR7614B (CASP Target)
To be Published
|
|
4DEP
 
 | | Structure of the IL-1b signaling complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-1 beta, Interleukin-1 receptor accessory protein, ... | | Authors: | Thomas, C, Garcia, K.C. | | Deposit date: | 2012-01-21 | | Release date: | 2012-03-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the activating IL-1 receptor signaling complex.
Nat.Struct.Mol.Biol., 19, 2012
|
|
