8J9H
 
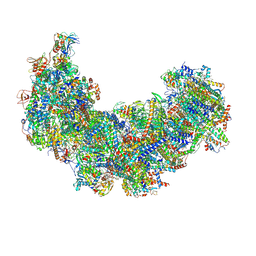 | | Cryo-EM structure of Euglena gracilis respiratory complex I, deactive state | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, CARDIOLIPIN, ... | | Authors: | Wu, M.C, He, Z.X, Tian, H.T, Hu, Y.Q, Han, F.Z, Zhou, L. | | Deposit date: | 2023-05-03 | | Release date: | 2024-02-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Euglena's atypical respiratory chain adapts to the discoidal cristae and flexible metabolism.
Nat Commun, 15, 2024
|
|
8J9G
 
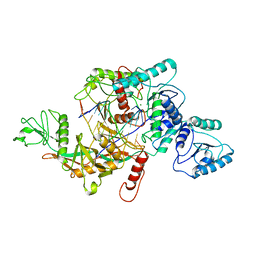 | | CrtSPARTA hetero-dimer bound with guide-target, state 1 | | Descriptor: | DNA (25-MER), MAGNESIUM ION, Piwi domain-containing protein, ... | | Authors: | Li, Z.X, Guo, L.J, Huang, P.P, Xiao, Y.B, Chen, M.R. | | Deposit date: | 2023-05-03 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Auto-inhibition and activation of a short Argonaute-associated TIR-APAZ defense system.
Nat.Chem.Biol., 20, 2024
|
|
8J9F
 
 | | Structure of STG-hydrolyzing beta-glucosidase 1 (PSTG1) | | Descriptor: | Beta-glucosidase, GLYCEROL | | Authors: | Yanai, T, Imaizumi, R, Takahashi, Y, Katsumura, E, Yamamoto, M, Nakayama, T, Yamashita, S, Takeshita, K, Sakai, N, Matsuura, H. | | Deposit date: | 2023-05-03 | | Release date: | 2024-04-10 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural insights into a bacterial beta-glucosidase capable of degrading sesaminol triglucoside to produce sesaminol: toward the understanding of the aglycone recognition mechanism by the C-terminal lid domain.
J.Biochem., 174, 2023
|
|
8J9D
 
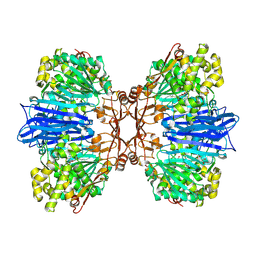 | | Crystal structure of M61 peptidase (bestatin-bound) from Xanthomonas campestris | | Descriptor: | 2-(3-AMINO-2-HYDROXY-4-PHENYL-BUTYRYLAMINO)-4-METHYL-PENTANOIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, ... | | Authors: | Yadav, P, Kumar, A, Kulkarni, B.S, Jamdar, S.N, Makde, R.D. | | Deposit date: | 2023-05-03 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a newly identified M61 family aminopeptidase with broad substrate specificity that is solely responsible for recycling acidic amino acids.
Febs J., 291, 2024
|
|
8J9C
 
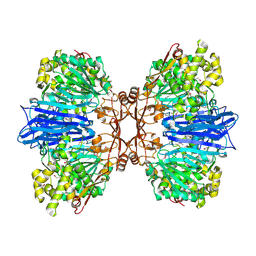 | | Crystal structure of M61 peptidase (apo-form) from Xanthomonas campestris | | Descriptor: | GLYCEROL, Putative glycyl aminopeptidase, SODIUM ION, ... | | Authors: | Yadav, P, Kumar, A, Jamdar, S.N, Makde, R.D. | | Deposit date: | 2023-05-03 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a newly identified M61 family aminopeptidase with broad substrate specificity that is solely responsible for recycling acidic amino acids.
Febs J., 291, 2024
|
|
8J99
 
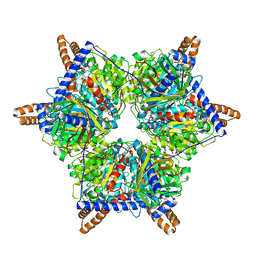 | | Human 3-methylcrotonyl-CoA carboxylase in BCS-mcoa state | | Descriptor: | Methylcrotonoyl-CoA carboxylase beta chain, mitochondrial, Methylcrotonoyl-CoA carboxylase subunit alpha, ... | | Authors: | Liu, D.S, Su, J.Y. | | Deposit date: | 2023-05-02 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Human 3-methylcrotonyl-CoA carboxylase in BCS-mcoa state
To Be Published
|
|
8J98
 
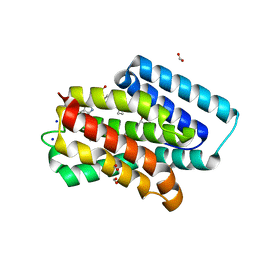 | | A near-infrared fluorescent protein of de novo backbone design | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethenyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, FORMIC ACID, GLYCEROL, ... | | Authors: | Hu, X, Xu, Y. | | Deposit date: | 2023-05-02 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | De novo backbone design for monomerization of near-infrared fluorescent protein
To Be Published
|
|
8J92
 
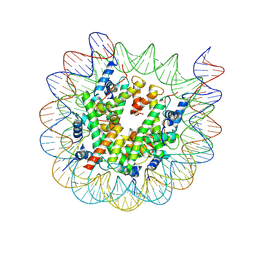 | | Cryo-EM structure of nucleosome containing Arabidopsis thaliana H2A.W | | Descriptor: | DNA (169-MER), HTA6, HTB9, ... | | Authors: | Osakabe, A, Takizawa, Y, Horikoshi, N, Hatazawa, S, Berger, F, Kurumizaka, H, Kakutani, T. | | Deposit date: | 2023-05-02 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular and structural basis of the chromatin remodeling activity by Arabidopsis DDM1.
Nat Commun, 15, 2024
|
|
8J91
 
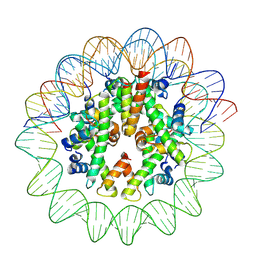 | | Cryo-EM structure of nucleosome containing Arabidopsis thaliana histones | | Descriptor: | DNA (169-MER), HTA13, Histone H2B.6, ... | | Authors: | Osakabe, A, Takizawa, Y, Horikoshi, N, Hatazawa, S, Berger, F, Kurumizaka, H, Kakutani, T. | | Deposit date: | 2023-05-02 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular and structural basis of the chromatin remodeling activity by Arabidopsis DDM1.
Nat Commun, 15, 2024
|
|
8J90
 
 | | Cryo-EM structure of DDM1-nucleosome complex | | Descriptor: | ATP-dependent DNA helicase DDM1, DNA (169-MER), HTA6, ... | | Authors: | Osakabe, A, Takizawa, Y, Horikoshi, N, Hatazawa, S, Berger, F, Kurumizaka, H, Kakutani, T. | | Deposit date: | 2023-05-02 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.71 Å) | | Cite: | Molecular and structural basis of the chromatin remodeling activity by Arabidopsis DDM1.
Nat Commun, 15, 2024
|
|
8J8X
 
 | | Crystal structure of AtHPPD-YH20307 complex | | Descriptor: | 4-hydroxyphenylpyruvate dioxygenase, 5-(1,3-dimethyl-5-oxidanyl-pyrazol-4-yl)carbonyl-1,4-dimethyl-3-propan-2-yl-benzimidazol-2-one, COBALT (II) ION | | Authors: | Lin, H.-Y, Dong, J, Yang, G.-F. | | Deposit date: | 2023-05-02 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | Crystal structure of AtHPPD-YH20307 complex
To Be Published
|
|
8J8T
 
 | |
8J8O
 
 | | Structure of Acb2 complexed with 2',3'-cGAMP | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, cGAMP, ... | | Authors: | Cao, X.L, Xiao, Y, Feng, Y. | | Deposit date: | 2023-05-02 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Phage anti-CBASS protein simultaneously sequesters cyclic trinucleotides and dinucleotides.
Mol.Cell, 84, 2024
|
|
8J8M
 
 | | Overall structure of the LAT1-4F2hc bound with tryptophan | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4F2 cell-surface antigen heavy chain, Large neutral amino acids transporter small subunit 1, ... | | Authors: | Yan, R.H, Li, Y.N, Shi, T.H. | | Deposit date: | 2023-05-02 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Overall structure of the LAT1-4F2hc bound with tryptophan
To Be Published
|
|
8J8H
 
 | | SPARTA monomer bound with guide-target, state 2 | | Descriptor: | DNA (25-MER), MAGNESIUM ION, Piwi domain-containing protein, ... | | Authors: | Li, Z.X, Guo, L.J, Huang, P.P, Xiao, Y.B, Chen, M.R. | | Deposit date: | 2023-05-01 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Auto-inhibition and activation of a short Argonaute-associated TIR-APAZ defense system.
Nat.Chem.Biol., 20, 2024
|
|
8J8G
 
 | | Monkeypox virus DNA replication holoenzyme F8, A22 and E4 in complex with a DNA duplex and cidofovir diphosphate | | Descriptor: | CALCIUM ION, DNA (5'-D(P*AP*GP*CP*TP*GP*CP*TP*AP*TP*GP*AP*GP*AP*TP*TP*AP*AP*GP*TP*TP*AP*T)-3'), DNA (5'-D(P*GP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*GP*AP*TP*AP*AP*CP*TP*TP*AP*AP*TP*CP*TP*CP*AP*CP*AP*TP*AP*GP*CP*AP*GP*CP*T)-3'), ... | | Authors: | Xu, Y, Wu, Y, Wu, X, Zhang, Y, Yang, Y, Li, D, Yang, B, Gao, K, Zhang, Z, Dong, C. | | Deposit date: | 2023-05-01 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Structural basis of human mpox viral DNA replication inhibition by brincidofovir and cidofovir.
Int.J.Biol.Macromol., 270, 2024
|
|
8J8D
 
 | |
8J8C
 
 | |
8J8B
 
 | |
8J8A
 
 | |
8J89
 
 | |
8J88
 
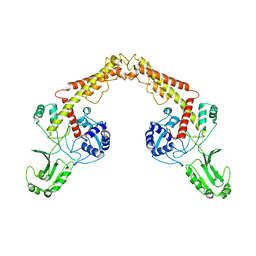 | | Asfv topoisomerase 2 - apo conformer Ib | | Descriptor: | DNA topoisomerase 2 | | Authors: | Chang, C.-W, Tsai, M.-D. | | Deposit date: | 2023-05-01 | | Release date: | 2024-02-07 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | A unified view on enzyme catalysis by cryo-EM study of a DNA topoisomerase.
Commun Chem, 7, 2024
|
|
8J87
 
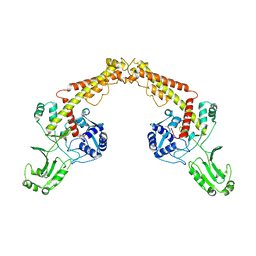 | | Asfv topoisomerase 2 - apo conformer Ia | | Descriptor: | DNA topoisomerase 2 | | Authors: | Chang, C.-W, Tsai, M.-D. | | Deposit date: | 2023-05-01 | | Release date: | 2024-02-07 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | A unified view on enzyme catalysis by cryo-EM study of a DNA topoisomerase.
Commun Chem, 7, 2024
|
|
8J85
 
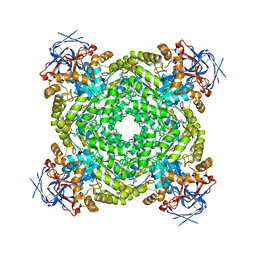 | | Cryo-EM structure of ochratoxin A-detoxifying amidohydrolase ADH3 mutant S88E in complex with ochratoxin A | | Descriptor: | (2~{S})-2-[[(3~{R})-5-chloranyl-3-methyl-8-oxidanyl-1-oxidanylidene-3,4-dihydroisochromen-7-yl]carbonylamino]-3-phenyl-propanoic acid, Amidohydrolase family protein, ZINC ION | | Authors: | Dai, L.H, Niu, D, Huang, J.-W, Li, X, Shen, P.P, Li, H, Hu, Y.M, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-04-30 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structure and rational engineering of a superefficient ochratoxin A-detoxifying amidohydrolase.
J Hazard Mater, 458, 2023
|
|
8J84
 
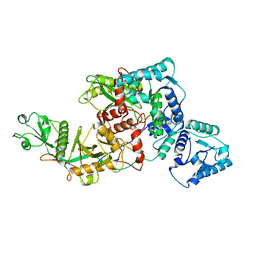 | | Short ago complexed with TIR-APAZ | | Descriptor: | Piwi domain-containing protein, TIR domain-containing protein | | Authors: | Guo, L.J, Huang, P.P, Li, Z.X, Xiao, Y.B, Chen, M.R. | | Deposit date: | 2023-04-30 | | Release date: | 2024-03-13 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Auto-inhibition and activation of a short Argonaute-associated TIR-APAZ defense system.
Nat.Chem.Biol., 20, 2024
|
|
