3I09
 
 | |
2YYL
 
 | | Crystal structure of the mutant of HpaB (T198I, A276G, and R466H) complexed with FAD | | Descriptor: | 4-hydroxyphenylacetate-3-hydroxylase, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Kim, S.-H, Hisano, T, Takeda, K, Iwasaki, W, Ebihara, A, Miki, K. | | Deposit date: | 2007-04-30 | | Release date: | 2007-09-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of the Oxygenase Component (HpaB) of the 4-Hydroxyphenylacetate 3-Monooxygenase from Thermus thermophilus HB8
J.Biol.Chem., 282, 2007
|
|
7ME0
 
 | | Cryo-EM structure of SARS-CoV-2 NSP15 NendoU at pH 6.0 | | Descriptor: | Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Song, Y, Nakamura, A.M, Noske, G.D, Gawriljuk, V.O, Fernandes, R.S, Oliva, G. | | Deposit date: | 2021-04-06 | | Release date: | 2021-04-14 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
2FNP
 
 | | Crystal structure of SarA | | Descriptor: | Staphylococcal accessory regulator A | | Authors: | Liu, Y, Manna, A.C, Pan, C.H, Cheung, A.L, Zhang, G. | | Deposit date: | 2006-01-11 | | Release date: | 2006-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and function analyses of the global regulatory protein SarA from Staphylococcus aureus.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
7M6B
 
 | | The Crystal Structure of Mcbe1 | | Descriptor: | 1,2-ETHANEDIOL, S-ADENOSYL-L-HOMOCYSTEINE, S-ADENOSYLMETHIONINE, ... | | Authors: | Alahuhta, P.M, Lunin, V.V. | | Deposit date: | 2021-03-25 | | Release date: | 2021-04-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Target highlights in CASP14: Analysis of models by structure providers.
Proteins, 89, 2021
|
|
5MQI
 
 | |
5MU2
 
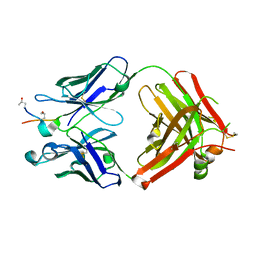 | | ACC1 Fab fragment in complex with CII583-591 (CG10) | | Descriptor: | ACC1 Fab fragment heavy chain, ACC1 Fab fragment light chain, synthetic peptide containing the CII583-591 epitope of collagen type II | | Authors: | Dobritzsch, D, Holmdahl, R, Ge, C. | | Deposit date: | 2017-01-12 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Anti-citrullinated protein antibodies cause arthritis by cross-reactivity to joint cartilage.
JCI Insight, 2, 2017
|
|
2PHH
 
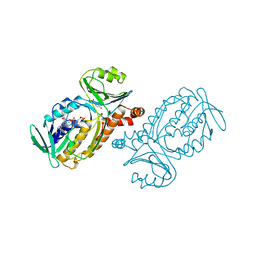 | |
5MWF
 
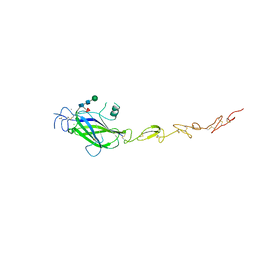 | | Human Jagged2 C2-EGF2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Suckling, R.J, Handford, P.A, Lea, S.M. | | Deposit date: | 2017-01-18 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and functional dissection of the interplay between lipid and Notch binding by human Notch ligands.
EMBO J., 36, 2017
|
|
7MFO
 
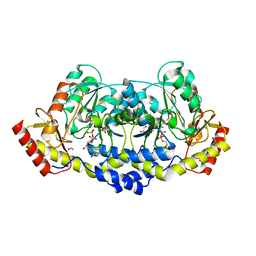 | | X-ray structure of the L136 Aminotransferase from Acanthamoeba polyphaga mimivirus in the presence of TDP and PMP | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, CHLORIDE ION, ... | | Authors: | Ferek, J.D, Thoden, J.B, Holden, H.M. | | Deposit date: | 2021-04-10 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Characterization of an aminotransferase from Acanthamoeba polyphaga Mimivirus.
Protein Sci., 30, 2021
|
|
2PHZ
 
 | | Crystal structure of Iron-uptake system-binding protein FeuA from Bacillus subtilis. Northeast Structural Genomics target SR580. | | Descriptor: | Iron-uptake system-binding protein | | Authors: | Benach, J, Neely, H, Seetharaman, J, Chen, C.X, Cunningham, K, Ma, L.-C, Janjua, H, Xiao, R, Baran, M, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-04-12 | | Release date: | 2007-04-24 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of Iron-uptake system-binding protein FeuA from Bacillus subtilis.
To be Published
|
|
5MXC
 
 | | Aleuria aurantia lectin (AAL) N224Q mutant in complex with alpha-methyl-L-fucoside | | Descriptor: | Fucose-specific lectin, GLYCEROL, methyl alpha-L-fucopyranoside | | Authors: | Houser, J, Kozmon, S, Romano, P.R, Wimmerova, M. | | Deposit date: | 2017-01-22 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Influence of Trp flipping on carbohydrate binding in lectins. An example on Aleuria aurantia lectin AAL.
PLoS ONE, 12, 2017
|
|
3I39
 
 | | NI,FE-CODH-320 MV+CN state | | Descriptor: | CYANIDE ION, Carbon monoxide dehydrogenase 2, FE (II) ION, ... | | Authors: | Jeoung, J.-H, Dobbek, H. | | Deposit date: | 2009-06-30 | | Release date: | 2009-08-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structural basis of cyanide inhibition of Ni, Fe-containing carbon monoxide dehydrogenase
J.Am.Chem.Soc., 131, 2009
|
|
7GE0
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with EDJ-MED-6af13d92-2 (Mpro-x11276) | | Descriptor: | 3C-like proteinase, 5-fluoro-N-[2-(2-methoxyphenoxy)ethyl]-2-oxo-1,2-dihydroquinoline-4-carboxamide, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GE2
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with EDJ-MED-6af13d92-1 (Mpro-x11313) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-[2-(2-methoxyphenoxy)ethyl]-5-methyl-2-oxo-1,2-dihydroquinoline-4-carboxamide | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
5MKF
 
 | | cryoEM Structure of Polycystin-2 in complex with calcium and lipids | | Descriptor: | 1,2-DIPALMITOYL-SN-GLYCERO-3-PHOSPHATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wilkes, M, Madej, M.G, Ziegler, C. | | Deposit date: | 2016-12-04 | | Release date: | 2017-01-18 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Molecular insights into lipid-assisted Ca(2+) regulation of the TRP channel Polycystin-2.
Nat. Struct. Mol. Biol., 24, 2017
|
|
2Y9J
 
 | |
2G3N
 
 | | Crystal structure of the Sulfolobus solfataricus alpha-glucosidase MalA in complex with beta-octyl-glucopyranoside | | Descriptor: | Alpha-glucosidase, octyl beta-D-glucopyranoside | | Authors: | Ernst, H.A, Lo Leggio, L, Willemoes, M, Leonard, G, Blum, P, Larsen, S. | | Deposit date: | 2006-02-20 | | Release date: | 2006-05-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of the Sulfolobus solfataricus alpha-Glucosidase: Implications for Domain Conservation and Substrate Recognition in GH31.
J.Mol.Biol., 358, 2006
|
|
3I5J
 
 | | Diferric Resting State Toluene 4-Monooxygenase HD complex | | Descriptor: | FE (III) ION, Toluene-4-monooxygenase system protein A, Toluene-4-monooxygenase system protein B, ... | | Authors: | Bailey, L.J, Fox, B.G. | | Deposit date: | 2009-07-05 | | Release date: | 2009-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic and catalytic studies of the peroxide-shunt reaction in a diiron hydroxylase.
Biochemistry, 48, 2009
|
|
5MTT
 
 | | Maltodextrin binding protein MalE1 from L. casei BL23 bound to maltotetraose | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MalE1, ... | | Authors: | Homburg, C, Bommer, M, Wuttge, S, Hobe, C, Beck, S, Dobbek, H, Deutscher, J, Licht, A, Schneider, E. | | Deposit date: | 2017-01-10 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Inducer exclusion in Firmicutes: insights into the regulation of a carbohydrate ATP binding cassette transporter from Lactobacillus casei BL23 by the signal transducing protein P-Ser46-HPr.
Mol. Microbiol., 105, 2017
|
|
3HS9
 
 | | Intersectin 1-peptide-AP2 beta ear complex | | Descriptor: | AP-2 complex subunit beta-1, peptide from Intersectin-1, residues 841-851 | | Authors: | Vahedi-Faridi, A, Pechstein, A, Schaefer, J.G, Saenger, W, Haucke, V. | | Deposit date: | 2009-06-10 | | Release date: | 2010-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Regulation of synaptic vesicle recycling by complex formation between intersectin 1 and the clathrin adaptor complex AP2.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3I69
 
 | | Apo Glutathione Transferase A1-1 GIMF-helix mutant | | Descriptor: | GLUTATHIONE, Glutathione S-transferase A1 | | Authors: | Balogh, L.M, Le Trong, I, Stenkamp, R.E, Atkins, W.M. | | Deposit date: | 2009-07-06 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural analysis of a glutathione transferase A1-1 mutant tailored for high catalytic efficiency with toxic alkenals.
Biochemistry, 48, 2009
|
|
2YNB
 
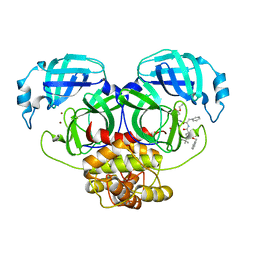 | |
2G6L
 
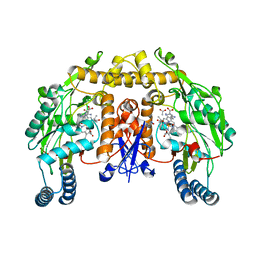 | | Structure of rat nNOS heme domain (BH2 bound) complexed with NO | | Descriptor: | 7,8-DIHYDROBIOPTERIN, ACETATE ION, ARGININE, ... | | Authors: | Li, H, Igarashi, J, Jamal, J, Yang, W, Poulos, T.L. | | Deposit date: | 2006-02-24 | | Release date: | 2006-08-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural studies of constitutive nitric oxide synthases with diatomic ligands bound.
J.Biol.Inorg.Chem., 11, 2006
|
|
5MO5
 
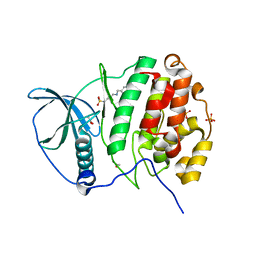 | | Crystal Structure of CK2alpha with N-(3-(((2-chloro-[1,1'-biphenyl]-4-yl)methyl)amino)propyl)methanesulfonamide bound | | Descriptor: | ACETATE ION, Casein kinase II subunit alpha, PHOSPHATE ION, ... | | Authors: | Brear, P, De Fusco, C, Georgiou, K, Iegre, J, Sore, H, Hyvonen, M, Spring, D. | | Deposit date: | 2016-12-13 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | A fragment-based approach leading to the discovery of a novel binding site and the selective CK2 inhibitor CAM4066.
Bioorg. Med. Chem., 25, 2017
|
|
