3MJY
 
 | | Crystal structure of dihydroorotate dehydrogenase from Leishmania major in complex with 5-Aminoorotic acid | | Descriptor: | 5-amino-2,6-dioxo-1,2,3,6-tetrahydropyrimidine-4-carboxylic acid, Dihydroorotate dehydrogenase, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Pinheiro, M.P, Rocha, J.R, Cheleski, J, Montanari, C.A, Nonato, M.C. | | Deposit date: | 2010-04-13 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Novel insights for dihydroorotate dehydrogenase class 1A inhibitors discovery.
Eur.J.Med.Chem., 45, 2010
|
|
3MHU
 
 | | Crystal structure of dihydroorotate dehydrogenase from Leishmania major in complex with 5-Nitroorotic acid | | Descriptor: | 5-nitro-2,6-dioxo-1,2,3,6-tetrahydropyrimidine-4-carboxylic acid, Dihydroorotate dehydrogenase, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Pinheiro, M.P, Rocha, J.R, Cheleski, J, Montanari, C.A, Nonato, M.C. | | Deposit date: | 2010-04-08 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Novel insights for dihydroorotate dehydrogenase class 1A inhibitors discovery.
Eur.J.Med.Chem., 45, 2010
|
|
5W4K
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with Klebsazolicin and bound to mRNA and A-, P- and E-site tRNAs at 2.7A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Metelev, M, Osterman, I.A, Ghilarov, D, Khabibullina, N.F, Yakimov, A, Shabalin, K, Utkina, I, Travin, D.Y, Komarova, E.S, Serebryakova, M, Artamonova, T, Khodorkovskii, M, Konevega, A.L, Sergiev, P.V, Severinov, K, Polikanov, Y.S. | | Deposit date: | 2017-06-12 | | Release date: | 2017-08-30 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Klebsazolicin inhibits 70S ribosome by obstructing the peptide exit tunnel.
Nat. Chem. Biol., 13, 2017
|
|
5VP2
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with madumycin II and bound to mRNA and A-, P- and E-site tRNAs at 2.8A resolution | | Descriptor: | 16S ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Osterman, I.A, Khabibullina, N.F, Komarova, E.S, Kasatsky, P, Kartsev, V.G, Bogdanov, A.A, Dontsova, O.A, Konevega, A.L, Sergiev, P.V, Polikanov, Y.S. | | Deposit date: | 2017-05-04 | | Release date: | 2017-06-28 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Madumycin II inhibits peptide bond formation by forcing the peptidyl transferase center into an inactive state.
Nucleic Acids Res., 45, 2017
|
|
5WF0
 
 | |
5WFK
 
 | |
5WFS
 
 | |
5WDT
 
 | | 70S ribosome-EF-Tu H84A complex with GppNHp | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Fislage, M, Brown, Z, Frank, J. | | Deposit date: | 2017-07-06 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM shows stages of initial codon selection on the ribosome by aa-tRNA in ternary complex with GTP and the GTPase-deficient EF-TuH84A.
Nucleic Acids Res., 46, 2018
|
|
5Y6P
 
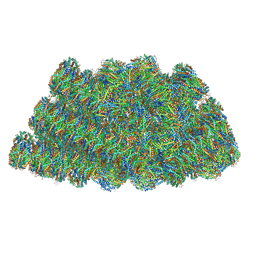 | | Structure of the phycobilisome from the red alga Griffithsia pacifica | | Descriptor: | ApcD, ApcF, LC, ... | | Authors: | Zhang, J, Ma, J.F, Liu, D.S, Sun, S, Sui, S.F. | | Deposit date: | 2017-08-13 | | Release date: | 2017-11-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of phycobilisome from the red alga Griffithsia pacifica
Nature, 551, 2017
|
|
5WE4
 
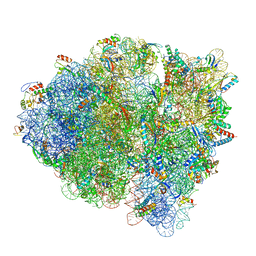 | | 70S ribosome-EF-Tu wt complex with GppNHp | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Fislage, M, Brown, Z, Frank, J. | | Deposit date: | 2017-07-07 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM shows stages of initial codon selection on the ribosome by aa-tRNA in ternary complex with GTP and the GTPase-deficient EF-TuH84A.
Nucleic Acids Res., 46, 2018
|
|
7YE3
 
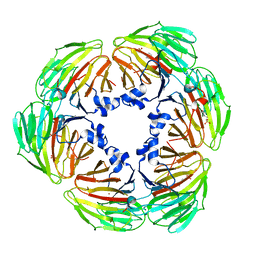 | | Crystal structure of Lactobacillus rhamnosus 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase KduI complexed with MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase, ZINC ION | | Authors: | Yamamoto, Y, Oiki, S, Takase, R, Mikami, B, Hashimoto, W. | | Deposit date: | 2022-07-05 | | Release date: | 2023-07-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.553 Å) | | Cite: | Crystal Structures of Lacticaseibacillus 4-Deoxy-L- threo- 5-hexosulose-uronate Ketol-isomerase KduI in Complex with Substrate Analogs.
J Appl Glycosci (1999), 70, 2023
|
|
3N9J
 
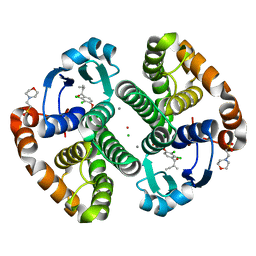 | |
6QE2
 
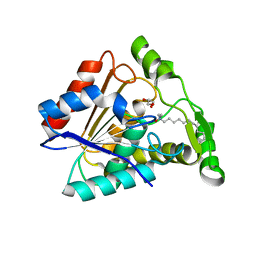 | | Crystal structure of Paleococcus ferrophilus monoacylglycerol lipase. | | Descriptor: | GLYCEROL, LAURYL DIMETHYLAMINE-N-OXIDE, Monoacylglycerol lipase | | Authors: | Labar, G, Demarez, M, Brandt, N, Wouters, J, Leherte, F. | | Deposit date: | 2019-01-04 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7461 Å) | | Cite: | Structure and Dynamics of an Archeal Monoglyceride Lipase from Palaeococcus ferrophilus as Revealed by Crystallography and In Silico Analysis.
Biomolecules, 11, 2021
|
|
4WDX
 
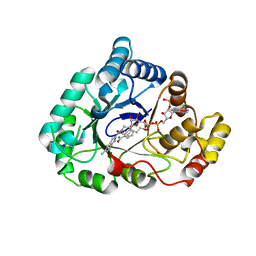 | | 17beta-HSD5 in complex with [4-(2-hydroxyethyl)piperidin-1-yl](5-methyl-1H-indol-2-yl)methanone | | Descriptor: | Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, [4-(2-hydroxyethyl)piperidin-1-yl](5-methyl-1H-indol-2-yl)methanone | | Authors: | Amano, Y, Yamaguchi, T. | | Deposit date: | 2014-09-09 | | Release date: | 2015-04-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structures of complexes of type 5 17 beta-hydroxysteroid dehydrogenase with structurally diverse inhibitors: insights into the conformational changes upon inhibitor binding.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5ZE0
 
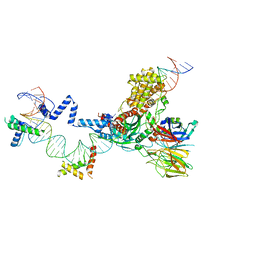 | | Hairpin Forming Complex, RAG1/2-Nicked(with Dideoxy) 12RSS/23RSS complex in Mg2+ | | Descriptor: | 1,2-ETHANEDIOL, DNA (30-MER), DNA (39-MER), ... | | Authors: | Kim, M.S, Chuenchor, W, Chen, X, Gellert, M, Yang, W. | | Deposit date: | 2018-02-25 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Cracking the DNA Code for V(D)J Recombination
Mol. Cell, 70, 2018
|
|
5ZK3
 
 | | Crystal structure of rationally thermostabilized M2 muscarinic acetylcholine receptor bound with QNB | | Descriptor: | (3R)-1-azabicyclo[2.2.2]oct-3-yl hydroxy(diphenyl)acetate, Muscarinic acetylcholine receptor M2,Apo-cytochrome b562,Muscarinic acetylcholine receptor M2 | | Authors: | Suno, R, Maeda, S, Yasuda, S, Yamashita, K, Hirata, K, Horita, S, Tawaramoto, M.S, Tsujimoto, H, Murata, T, Kinoshita, M, Yamamoto, M, Kobilka, B.K, Iwata, S, Kobayashi, T. | | Deposit date: | 2018-03-23 | | Release date: | 2018-11-21 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into the subtype-selective antagonist binding to the M2muscarinic receptor
Nat. Chem. Biol., 14, 2018
|
|
1S38
 
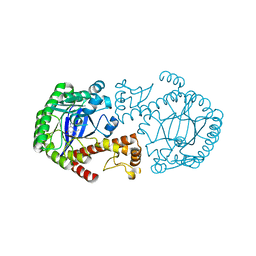 | | CRYSTAL STRUCTURE OF TGT IN COMPLEX WITH 2-AMINO-8-METHYLQUINAZOLIN-4(3H)-ONE | | Descriptor: | 2-AMINO-8-METHYLQUINAZOLIN-4(3H)-ONE, ZINC ION, tRNA guanine transglycosylase | | Authors: | Meyer, E.A, Furler, M, Diederich, F, Brenk, R, Klebe, G. | | Deposit date: | 2004-01-12 | | Release date: | 2004-07-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Synthesis and In vitro Evaluation of 2-Aminoquinazolin-4(3H)-one-based Inhibitors for tRNA-Guanine Transglycosylase (TGT)
HELV.CHIM.ACTA, 87, 2004
|
|
4U5Y
 
 | | Crystal Structure of the complex between the GNAT domain of S. lividans PAT and the acetyl-CoA synthetase C-terminal domain of S. enterica | | Descriptor: | Acetyl-coenzyme A synthetase, Acyl-CoA synthetase, SULFATE ION | | Authors: | Taylor, K.C, Tucker, A.C, Rank, K.C, Escalante-Semerena, J.C, Rayment, I. | | Deposit date: | 2014-07-25 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.926 Å) | | Cite: | Insights into the specificity of lysine acetyltransferases.
J.Biol.Chem., 289, 2014
|
|
5Z99
 
 | |
5ZE1
 
 | | Hairpin Forming Complex, RAG1/2-Nicked 12RSS/23RSS complex in 2mM Mn2+ for 10 min at 4'C | | Descriptor: | 1,2-ETHANEDIOL, DNA, HMGB1 A-B box, ... | | Authors: | Kim, M.S, Chuenchor, W, Chen, X, Gellert, M, Yang, W. | | Deposit date: | 2018-02-25 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Cracking the DNA Code for V(D)J Recombination
Mol. Cell, 70, 2018
|
|
5ZFJ
 
 | | Crystal structure of a cyclase Filc from Fischerella sp. in complex with 4-(1H-Indol-3-yl)butan-2-one | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(1~{H}-indol-3-yl)butan-2-one, CALCIUM ION, ... | | Authors: | Hu, X.Y, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-03-06 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The Crystal Structure of a Class of Cyclases that Catalyze the Cope Rearrangement
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6Q0N
 
 | | Structure of the Erbin PDB domain in complex with a high-affinity peptide | | Descriptor: | Erbin, peptide | | Authors: | Singer, A.U, Teyra, J, Ernst, A, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2019-08-02 | | Release date: | 2019-11-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Comprehensive analysis of all evolutionary paths between two divergent PDZ domain specificities.
Protein Sci., 29, 2020
|
|
6Q0M
 
 | | Structure of Erbin PDZ derivative E-14 with a high-affinity peptide | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Singer, A.U, Teyra, J, Ernst, A, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2019-08-02 | | Release date: | 2019-11-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Comprehensive analysis of all evolutionary paths between two divergent PDZ domain specificities.
Protein Sci., 29, 2020
|
|
3QKG
 
 | |
4WDW
 
 | | 17beta-HSD5 in complex with 3,6-dihydropyridin-1(2H)-yl(5-methyl-1H-indol-2-yl)methanone | | Descriptor: | 3,6-dihydropyridin-1(2H)-yl(5-methyl-1H-indol-2-yl)methanone, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Amano, Y, Yamaguchi, T. | | Deposit date: | 2014-09-09 | | Release date: | 2015-04-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structures of complexes of type 5 17 beta-hydroxysteroid dehydrogenase with structurally diverse inhibitors: insights into the conformational changes upon inhibitor binding.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
