1W79
 
 | | Crystal structure of the DD-transpeptidase-carboxypeptidase from Actinomadura R39 | | Descriptor: | D-alanyl-D-alanine carboxypeptidase, MAGNESIUM ION, SULFATE ION | | Authors: | Sauvage, E, Herman, R, Petrella, S, Duez, C, Frere, J.M, Charlier, P. | | Deposit date: | 2004-08-31 | | Release date: | 2005-06-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the Actinomadura R39 DD-peptidase reveals new domains in penicillin-binding proteins.
J. Biol. Chem., 280, 2005
|
|
8TCJ
 
 | |
8TCM
 
 | | Crystal Structure of modified HIV reverse transcriptase p51 domain (FPC1) with picric acid and Xanthene-1,3,6,8-tetrol bound | | Descriptor: | 9H-xanthene-1,3,6,8-tetrol, PICRIC ACID, p51 subunit | | Authors: | Pedersen, L.C, London, R.E. | | Deposit date: | 2023-07-02 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal Structure of modified HIV reverse transcriptase p51 domain (FPC1) with picric acid and xanthene bound
To Be Published
|
|
8TIA
 
 | |
8TI8
 
 | |
8TI9
 
 | |
1GSV
 
 | | Crystal structure of the P65 crystal form of photoactive yellow protein G47S mutant | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | Authors: | Van Aalten, D.M.F, Crielaard, W, Hellingwerf, K.J, Joshua-Tor, L. | | Deposit date: | 2002-01-08 | | Release date: | 2002-02-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Engineering Photocycle Dynamics: Crystal Structures and Kinetics of Three Photoactive Yellow Protein Hinge-Bending Mutants
J.Biol.Chem., 227, 2002
|
|
8TCL
 
 | |
8A51
 
 | | Crystal structure of HSF2BP-BRME1 complex | | Descriptor: | 1,2-ETHANEDIOL, Break repair meiotic recombinase recruitment factor 1, CHLORIDE ION, ... | | Authors: | Miron, S, Legrand, P, Ropars, V, Ghouil, R, Zinn-Justin, S. | | Deposit date: | 2022-06-13 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | BRCA2-HSF2BP oligomeric ring disassembly by BRME1 promotes homologous recombination.
Sci Adv, 9, 2023
|
|
2W0X
 
 | | FACTOR INHIBITING HIF-1 ALPHA WITH PYRIDINE 2,4 DICARBOXYLIC ACID | | Descriptor: | FE (II) ION, GLYCEROL, HYPOXIA-INDUCIBLE FACTOR 1 ALPHA INHIBITOR, ... | | Authors: | Conejo-Garcia, A, Lienard, B.M.R, Clifton, I.J, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2008-10-10 | | Release date: | 2009-11-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural basis for binding of cyclic 2-oxoglutarate analogues to factor-inhibiting hypoxia-inducible factor.
Bioorg. Med. Chem. Lett., 20, 2010
|
|
1VPO
 
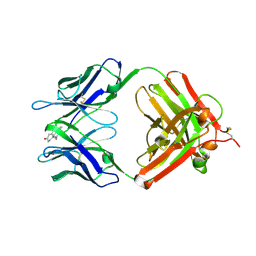 | | Crystal Structure Analysis of the Anti-testosterone Fab in Complex with Testosterone | | Descriptor: | TESTOSTERONE, anti-testosterone (heavy chain), anti-testosterone (light chain) | | Authors: | Valjakka, J.M, Hemminki, A, Niemi, S, Soderlund, H, Takkinen, K, Rouvinen, J. | | Deposit date: | 2004-11-15 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of an in Vitro Affinity- and Specificity-matured Anti-testosterone
Fab in Complex with Testosterone. IMPROVED AFFINITY RESULTS FROM
SMALL STRUCTURAL CHANGES WITHIN THE VARIABLE DOMAINS
J.Biol.Chem., 277, 2002
|
|
1VJ9
 
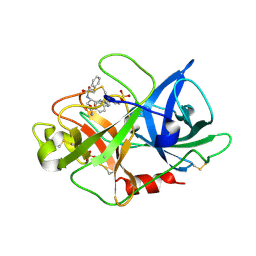 | | Urokinase Plasminogen Activator B-Chain-JT464 Complex | | Descriptor: | N-(BENZYLSULFONYL)-L-SERYL-N~1~-{4-[AMINO(IMINO)METHYL]BENZYL}-O-BENZYL-L-SERINAMIDE, SULFATE ION, plasminogen activator, ... | | Authors: | Schweinitz, A, Steinmetzer, T, Banke, I.J, Arlt, M.J.E, Stuerzebecher, A, Schuster, O, Geissler, A, Giersiefen, H, Zeslawska, E, Jacob, U, Kruger, A, Stuerzebecher, J. | | Deposit date: | 2004-02-03 | | Release date: | 2004-06-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design of novel and selective inhibitors of urokinase-type plasminogen activator with improved pharmacokinetic properties for use as antimetastatic agents
J.Biol.Chem., 279, 2004
|
|
1VQP
 
 | |
1H2K
 
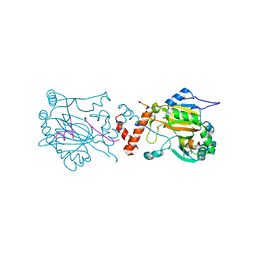 | | Factor Inhibiting HIF-1 alpha in complex with HIF-1 alpha fragment peptide | | Descriptor: | FACTOR INHIBITING HIF1, FE (II) ION, HYPOXIA-INDUCIBLE FACTOR 1 ALPHA, ... | | Authors: | Elkins, J.M, Hewitson, K.S, McNeill, L.A, Schlemminger, I, Seibel, J.F, Schofield, C.J. | | Deposit date: | 2002-08-12 | | Release date: | 2002-11-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of Factor-Inhibiting Hypoxia-Inducible Factor (Hif) Reveals Mechanism of Oxidative Modification of Hif-1Alpha
J.Biol.Chem., 278, 2003
|
|
2VPK
 
 | | Crystal structure of the BTB domain of human myoneurin | | Descriptor: | MYONEURIN | | Authors: | Cooper, C.D.O, Murray, J.W, Bullock, A, Pike, A.C.W, von Delft, F, Filippakopoulos, P, Salah, E, Edwards, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Knapp, S. | | Deposit date: | 2008-02-29 | | Release date: | 2008-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Btb Domain of Human Myoneurin
To be Published
|
|
8A8F
 
 | | Crystal structure of Glc7 phosphatase in complex with the regulatory region of Ref2 | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, RNA end formation protein 2, ... | | Authors: | Carminati, M, Manav, C.M, Bellini, D, Passmore, L.A. | | Deposit date: | 2022-06-22 | | Release date: | 2023-11-29 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A direct interaction between CPF and RNA Pol II links RNA 3' end processing to transcription.
Mol.Cell, 83, 2023
|
|
1H2M
 
 | | Factor Inhibiting HIF-1 alpha in complex with HIF-1 alpha fragment peptide | | Descriptor: | FACTOR INHIBITING HIF1, HYPOXIA-INDUCIBLE FACTOR 1 ALPHA, N-OXALYLGLYCINE, ... | | Authors: | Elkins, J.M, Hewitson, K.S, McNeill, L.A, Schlemminger, I, Seibel, J.F, Schofield, C.J. | | Deposit date: | 2002-08-12 | | Release date: | 2002-11-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Factor-Inhibiting Hypoxia-Inducible Factor (Hif) Reveals Mechanism of Oxidative Modification of Hif-1Alpha
J.Biol.Chem., 278, 2003
|
|
1W10
 
 | | Urokinase type plasminogen activator | | Descriptor: | N-(ISOBUTOXYCARBONYL)-D-SERYL-N-((1S)-4-{[AMINO(IMINO)METHYL]AMINO}-1-FORMYLBUTYL)-L-ALANINAMIDE, SULFATE ION, UROKINASE-TYPE PLASMINOGEN ACTIVATOR | | Authors: | Jacob, U. | | Deposit date: | 2004-06-15 | | Release date: | 2008-05-20 | | Last modified: | 2019-09-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystals of urokinase type plasminogen activator complexes reveal the binding mode of peptidomimetic inhibitors.
J.Mol.Biol., 328, 2003
|
|
2X0L
 
 | | Crystal structure of a neuro-specific splicing variant of human histone lysine demethylase LSD1. | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, HISTONE H3 PEPTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1, ... | | Authors: | Zibetti, C, Adamo, A, Binda, C, Forneris, F, Verpelli, C, Ginelli, E, Mattevi, A, Sala, C, Battaglioli, E. | | Deposit date: | 2009-12-15 | | Release date: | 2010-03-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Alternative Splicing of the Histone Demethylase Lsd1/Kdm1 Contributes to the Modulation of Neurite Morphogenesis in the Mammalian Nervous System.
J.Neurosci., 30, 2010
|
|
1VQM
 
 | |
1W12
 
 | | UROKINASE TYPE PLASMINOGEN ACTIVATOR | | Descriptor: | N-((1S)-4-{[AMINO(IMINO)METHYL]AMINO}-1-FORMYLBUTYL)-2-{(3R)-3-[(BENZYLSULFONYL)AMINO]-2-OXO-5-PHENYL-2,3-DIHYDRO-1H-1,4-BENZODIAZEPIN-1-YL}ACETAMIDE, UROKINASE-TYPE PLASMINOGEN ACTIVATOR | | Authors: | Jacob, U. | | Deposit date: | 2004-06-15 | | Release date: | 2008-05-20 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystals of Urokinase Type Plasminogen Activator Complexes Reveal the Binding Mode of Peptidomimetic Inhibitors.
J.Mol.Biol., 328, 2003
|
|
1VSO
 
 | | Crystal Structure of the Ligand-Binding Core of iGluR5 in Complex With the Antagonist (S)-ATPO at 1.85 A resolution | | Descriptor: | (S)-2-AMINO-3-(5-TERT-BUTYL-3-(PHOSPHONOMETHOXY)-4-ISOXAZOLYL)PROPIONIC ACID, GLYCEROL, Glutamate receptor, ... | | Authors: | Hald, H, Naur, P, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2007-03-29 | | Release date: | 2007-07-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Partial agonism and antagonism of the ionotropic glutamate receptor iGLuR5: structures of the ligand-binding core in complex with domoic acid and 2-amino-3-[5-tert-butyl-3-(phosphonomethoxy)-4-isoxazolyl]propionic acid.
J.Biol.Chem., 282, 2007
|
|
2X35
 
 | | Molecular basis of Histone H3K36me3 recognition by the PWWP domain of BRPF1. | | Descriptor: | PEREGRIN | | Authors: | Vezzoli, A, Bonadies, N, Allen, M.D, Freund, S.M.V, Santiveri, C.M, Kvinlaug, B, Huntly, B.J.P, Gottgens, B, Bycroft, M. | | Deposit date: | 2010-01-21 | | Release date: | 2010-02-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular Basis of Histone H3K36Me3 Recognition by the Pwwp Domain of Brpf1.
Nat.Struct.Mol.Biol., 17, 2010
|
|
1W0Z
 
 | | Urokinase type plasminogen activator | | Descriptor: | N-(BUTYLSULFONYL)-D-SERYL-N-{4-[AMINO(IMINO)METHYL]BENZYL}-L-ALANINAMIDE, SULFATE ION, UROKINASE-TYPE PLASMINOGEN ACTIVATOR | | Authors: | Jacob, U. | | Deposit date: | 2004-06-15 | | Release date: | 2008-05-20 | | Last modified: | 2019-09-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystals of urokinase type plasminogen activator complexes reveal the binding mode of peptidomimetic inhibitors.
J.Mol.Biol., 328, 2003
|
|
1VQ9
 
 | |
