4K2N
 
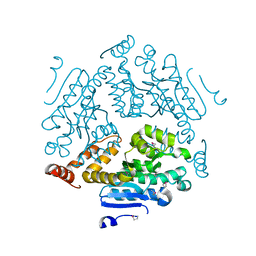 | | Crystal structure of an enoyl-CoA hydratase/ carnithine racemase from Magnetospirillum magneticum | | Descriptor: | Enoyl-CoA hydratase/carnithine racemase | | Authors: | Eswaramoorthy, S, Chamala, S, Evans, B, Foti, F, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Al Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-09 | | Release date: | 2013-04-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of an enoyl-CoA hydratase/ carnithine racemase from Magnetospirillum magneticum
To be Published
|
|
1DV6
 
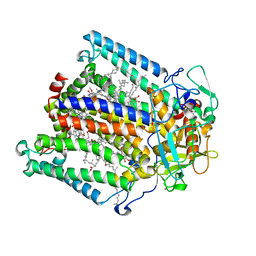 | | PHOTOSYNTHETIC REACTION CENTER FROM RHODOBACTER SPHAEROIDES IN THE CHARGE-NEUTRAL DQAQB STATE WITH THE PROTON TRANSFER INHIBITOR ZN2+ | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CHLORIDE ION, ... | | Authors: | Axelrod, H.L, Abresch, E.C, Paddock, M.L, Okamura, M.Y, Feher, G. | | Deposit date: | 2000-01-19 | | Release date: | 2000-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Determination of the binding sites of the proton transfer inhibitors Cd2+ and Zn2+ in bacterial reaction centers.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1E0C
 
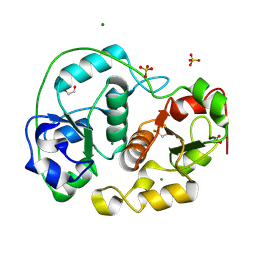 | | SULFURTRANSFERASE FROM AZOTOBACTER VINELANDII | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Bordo, D, Deriu, D, Colnaghi, R, Carpen, A, Pagani, S, Bolognesi, M. | | Deposit date: | 2000-03-23 | | Release date: | 2000-05-08 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Crystal Structure of a Sulfurtransferase from Azotobacter Vinelandii Highlights the Evolutionary Relationship between the Rhodanese and Phosphatase Enzyme Families
J.Mol.Biol., 298, 2000
|
|
1DST
 
 | |
7KHS
 
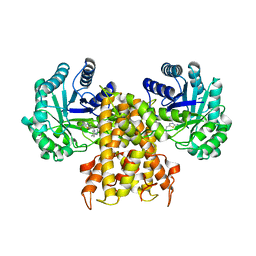 | | OgOGA IN COMPLEX WITH LIGAND 55 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, N-(5-{[(5S)-7-(5-methylimidazo[1,2-a]pyrimidin-7-yl)-2,7-diazaspiro[4.4]nonan-2-yl]methyl}-1,3-thiazol-2-yl)acetamide, ... | | Authors: | Shaffer, P.L. | | Deposit date: | 2020-10-21 | | Release date: | 2020-12-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Diazaspirononane Nonsaccharide Inhibitors of O-GlcNAcase (OGA) for the Treatment of Neurodegenerative Disorders.
J.Med.Chem., 63, 2020
|
|
1CLF
 
 | | CLOSTRIDIUM PASTEURIANUM FERREDOXIN | | Descriptor: | FERREDOXIN, IRON/SULFUR CLUSTER | | Authors: | Bertini, I, Donaire, A, Feinberg, B.A, Luchinat, C, Piccioli, M, Yuan, H. | | Deposit date: | 1995-06-21 | | Release date: | 1996-01-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the oxidized 2[4Fe-4S] ferredoxin from Clostridium pasteurianum.
Eur.J.Biochem., 232, 1995
|
|
4IRF
 
 | |
1CPR
 
 | | ST. LOUIS CYTOCHROME C' FROM THE PURPLE PHOTOTROPIC BACTERIUM, RHODOBACTER CAPSULATUS | | Descriptor: | CYTOCHROME C', PROTOPORPHYRIN IX CONTAINING FE, ZINC ION | | Authors: | Tahirov, T.H, Misaki, S, Meyer, T.E, Cusanovich, M.A, Yasuoka, N. | | Deposit date: | 1996-04-09 | | Release date: | 1997-01-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of cytochrome c' from Rhodobacter capsulatus strain St Louis: an unusual molecular association induced by bridging Zn ions.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
1OVD
 
 | | THE K136E MUTANT OF LACTOCOCCUS LACTIS DIHYDROOROTATE DEHYDROGENASE A IN COMPLEX WITH OROTATE | | Descriptor: | DIHYDROOROTATE DEHYDROGENASE A, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Norager, S, Arent, S, Bjornberg, O, Ottosen, M, Lo Leggio, L, Jensen, K.F, Larsen, S. | | Deposit date: | 2003-03-26 | | Release date: | 2003-08-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Lactococcus lactis dihydroorotate dehydrogenase A mutants reveal important facets of the enzymatic function.
J.Biol.Chem., 278, 2003
|
|
1L9U
 
 | | THERMUS AQUATICUS RNA POLYMERASE HOLOENZYME AT 4 A RESOLUTION | | Descriptor: | MAGNESIUM ION, RNA POLYMERASE, ALPHA SUBUNIT, ... | | Authors: | Murakami, K.S, Masuda, S, Darst, S.A. | | Deposit date: | 2002-03-26 | | Release date: | 2002-05-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural basis of transcription initiation: RNA polymerase holoenzyme at 4 A resolution.
Science, 296, 2002
|
|
4JCS
 
 | | Crystal structure of Enoyl-CoA hydratase/isomerase from Cupriavidus metallidurans CH34 | | Descriptor: | Enoyl-CoA hydratase/isomerase | | Authors: | Eswaramoorthy, S, Chamala, S, Chamala, B, Foti, F, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Al Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-02-22 | | Release date: | 2013-03-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of Enoyl-CoA hydratase/isomerase from Cupriavidus metallidurans CH34
To be Published
|
|
2CGA
 
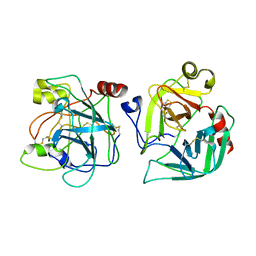 | |
1KZ1
 
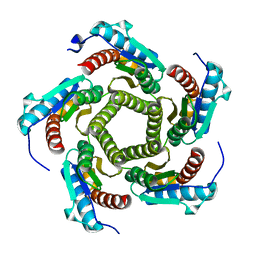 | | Mutant enzyme W27G Lumazine Synthase from S.pombe | | Descriptor: | 6,7-Dimethyl-8-ribityllumazine Synthase | | Authors: | Gerhardt, S, Haase, I, Steinbacher, S, Kaiser, J.T, Cushman, M, Bacher, A, Huber, R, Fischer, M. | | Deposit date: | 2002-02-06 | | Release date: | 2002-07-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structural basis of riboflavin binding to Schizosaccharomyces pombe 6,7-dimethyl-8-ribityllumazine synthase.
J.Mol.Biol., 318, 2002
|
|
1DSU
 
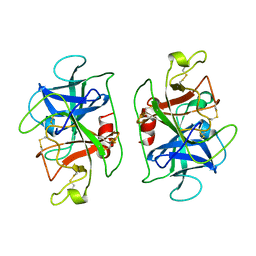 | |
3CR6
 
 | |
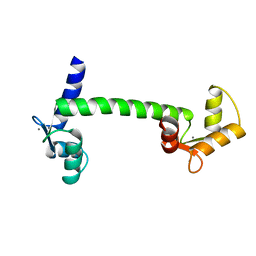
3CLN
 
 | |
2PV7
 
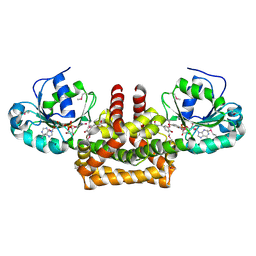 | |
2RS8
 
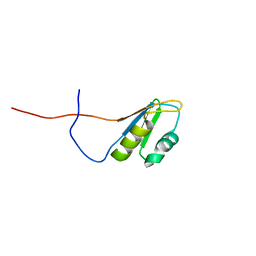 | | Solution structure of the N-terminal RNA recognition motif of NonO | | Descriptor: | Non-POU domain-containing octamer-binding protein | | Authors: | Nagata, T, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-11-29 | | Release date: | 2012-12-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal RNA recognition motif of NonO
To be Published
|
|
1DE3
 
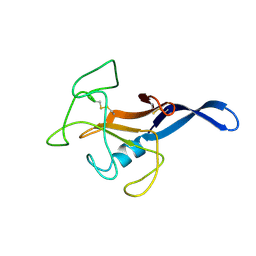 | | SOLUTION STRUCTURE OF THE CYTOTOXIC RIBONUCLEASE ALPHA-SARCIN | | Descriptor: | RIBONUCLEASE ALPHA-SARCIN | | Authors: | Perez-Canadillas, J.M, Campos-Olivas, R, Santoro, J, Lacadena, J, Martinez del Pozo, A, Gavilanes, J.G, Rico, M, Bruix, M. | | Deposit date: | 1999-11-12 | | Release date: | 2000-06-21 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | The highly refined solution structure of the cytotoxic ribonuclease alpha-sarcin reveals the structural requirements for substrate recognition and ribonucleolytic activity.
J.Mol.Biol., 299, 2000
|
|
2RCS
 
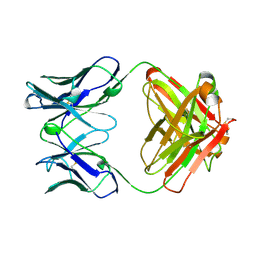 | | IMMUNOGLOBULIN 48G7 GERMLINE FAB-AFFINITY MATURATION OF AN ESTEROLYTIC ANTIBODY | | Descriptor: | IMMUNOGLOBULIN 48G7 GERMLINE FAB | | Authors: | Wedemayer, G.J, Wang, L.H, Patten, P.A, Schultz, P.G, Stevens, R.C. | | Deposit date: | 1997-05-14 | | Release date: | 1997-11-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into the evolution of an antibody combining site.
Science, 276, 1997
|
|
5UKR
 
 | |
1UUC
 
 | |
4JFC
 
 | | Crystal structure of a enoyl-CoA hydratase from Polaromonas sp. JS666 | | Descriptor: | Enoyl-CoA hydratase, GLYCEROL | | Authors: | Kumaran, D, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Al Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-02-28 | | Release date: | 2013-05-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of a enoyl-CoA hydratase from Polaromonas sp. JS666
To be Published
|
|
1ED1
 
 | | CRYSTAL STRUCTURE OF SIMIAN IMMUNODEFICIENCY VIRUS MATRIX ANTIGEN (SIV MA) AT 100K. | | Descriptor: | GAG POLYPROTEIN, ISOPROPYL ALCOHOL | | Authors: | Rao, Z, Belyaev, A, Fry, E, Roy, P, Jones, I.M, Stuart, D.I. | | Deposit date: | 2000-01-26 | | Release date: | 2000-02-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of SIV matrix antigen and implications for virus assembly.
Nature, 378, 1995
|
|
1Q0N
 
 | | CRYSTAL STRUCTURE OF A TERNARY COMPLEX OF 6-HYDROXYMETHYL-7,8-DIHYDROPTERIN PYROPHOSPHOKINASE FROM E. COLI WITH MGAMPCPP AND 6-HYDROXYMETHYL-7,8-DIHYDROPTERIN AT 1.25 ANGSTROM RESOLUTION | | Descriptor: | 2-AMINO-6-HYDROXYMETHYL-7,8-DIHYDRO-3H-PTERIDIN-4-ONE, 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, ACETATE ION, ... | | Authors: | Blaszczyk, J, Ji, X. | | Deposit date: | 2003-07-16 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Catalytic Center Assembly of Hppk as Revealed by the Crystal Structure of a Ternary Complex at 1.25 A Resolution
Structure, 8, 2000
|
|
