6ERG
 
 | | Complex of XLF and heterodimer Ku bound to DNA | | Descriptor: | DNA (21-MER), DNA (34-MER), Non-homologous end-joining factor 1, ... | | Authors: | Nemoz, C, Legrand, P, Ropars, V, Charbonnier, J.B. | | Deposit date: | 2017-10-18 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | XLF and APLF bind Ku80 at two remote sites to ensure DNA repair by non-homologous end joining.
Nat. Struct. Mol. Biol., 25, 2018
|
|
8IO4
 
 | | Herg1a-herg1b open state | | Descriptor: | Potassium voltage-gated channel subfamily H member 2 | | Authors: | Zhang, M.F. | | Deposit date: | 2023-03-10 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of Herg1a-herg1b open state
To Be Published
|
|
7YTB
 
 | | Crystal structure of Kin4B8 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Kin4B8, RETINAL | | Authors: | Murakoshi, S, Chazan, A, Shihoya, W, Beja, O, Nureki, O. | | Deposit date: | 2022-08-14 | | Release date: | 2023-03-15 | | Last modified: | 2023-03-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Phototrophy by antenna-containing rhodopsin pumps in aquatic environments.
Nature, 615, 2023
|
|
8I2Z
 
 | | Cryo-EM structure of the zeaxanthin-bound kin4B8 | | Descriptor: | RETINAL, Xanthorhodopsin, Zeaxanthin | | Authors: | Murakoshi, S, Chazan, A, Shihoya, W, Beja, O, Nureki, O. | | Deposit date: | 2023-01-15 | | Release date: | 2023-03-29 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | Phototrophy by antenna-containing rhodopsin pumps in aquatic environments.
Nature, 615, 2023
|
|
4U8I
 
 | | Structure of Aspergillus fumigatus UDP-Galactopyranose mutase mutant F66A | | Descriptor: | 1,2-ETHANEDIOL, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Qureshi, I.A, Chaudhary, R, Tanner, J.J. | | Deposit date: | 2014-08-03 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Contributions of Unique Active Site Residues of Eukaryotic UDP-Galactopyranose Mutases to Substrate Recognition and Active Site Dynamics.
Biochemistry, 53, 2014
|
|
4U8P
 
 | | Structure of Aspergillus fumigatus UDP-Galactopyranose mutase mutant Y317A complexed with UDP | | Descriptor: | 1,2-ETHANEDIOL, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Qureshi, I.A, Chaudhary, R, Tanner, J.J. | | Deposit date: | 2014-08-03 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Contributions of Unique Active Site Residues of Eukaryotic UDP-Galactopyranose Mutases to Substrate Recognition and Active Site Dynamics.
Biochemistry, 53, 2014
|
|
4U8O
 
 | | Structure of Aspergillus fumigatus UDP-Galactopyranose mutase mutant N207A complexed with UDP | | Descriptor: | 1,2-ETHANEDIOL, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Qureshi, I.A, Chaudhary, R, Tanner, J.J. | | Deposit date: | 2014-08-03 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Contributions of Unique Active Site Residues of Eukaryotic UDP-Galactopyranose Mutases to Substrate Recognition and Active Site Dynamics.
Biochemistry, 53, 2014
|
|
4KRP
 
 | |
4KRL
 
 | |
4KRM
 
 | |
4TVJ
 
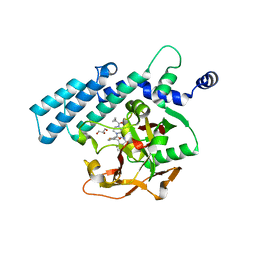 | | HUMAN ARTD2 (PARP2) - CATALYTIC DOMAIN IN COMPLEX WITH OLAPARIB | | Descriptor: | 4-(3-{[4-(cyclopropylcarbonyl)piperazin-1-yl]carbonyl}-4-fluorobenzyl)phthalazin-1(2H)-one, GLYCEROL, Poly [ADP-ribose] polymerase 2 | | Authors: | Karlberg, T, Thorsell, A.G, Ekblad, T, Pinto, A.F, Schuler, H. | | Deposit date: | 2014-06-27 | | Release date: | 2015-07-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Potency and Promiscuity in Poly(ADP-ribose) Polymerase (PARP) and Tankyrase Inhibitors.
J. Med. Chem., 60, 2017
|
|
4TZL
 
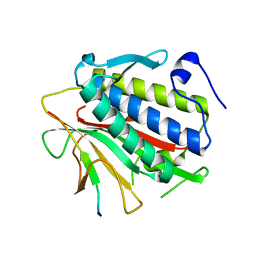 | |
4TZS
 
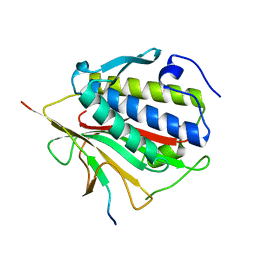 | |
4TZJ
 
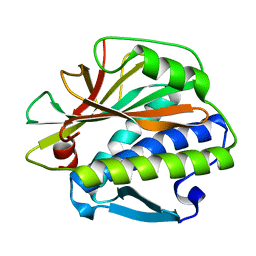 | |
4TZM
 
 | |
7OWL
 
 | | Odinarchaeota Adenylate kinase (OdinAK) in complex with CTP | | Descriptor: | Adenylate kinase, CYTIDINE-5'-TRIPHOSPHATE | | Authors: | Aberg-Zingmark, E, Grundstrom, C, Verma, A, Wolf-Watz, M, Sauer, U.H, Sauer-Eriksson, A.E. | | Deposit date: | 2021-06-18 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Insights into the evolution of enzymatic specificity and catalysis: From Asgard archaea to human adenylate kinases.
Sci Adv, 8, 2022
|
|
7OWK
 
 | | Odinarchaeota Adenylate kinase (OdinAK) in complex with dTTP | | Descriptor: | Adenylate kinase, THYMIDINE-5'-TRIPHOSPHATE | | Authors: | Aberg-Zingmark, E, Grundstrom, C, Verma, A, Wolf-Watz, M, Sauer, U.H, Sauer-Eriksson, A.E. | | Deposit date: | 2021-06-18 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Insights into the evolution of enzymatic specificity and catalysis: From Asgard archaea to human adenylate kinases.
Sci Adv, 8, 2022
|
|
7OWH
 
 | | Odinarchaeota Adenylate kinase (OdinAK) native structure | | Descriptor: | Adenylate kinase, CHLORIDE ION | | Authors: | Aberg-Zingmark, E, Grundstrom, C, Verma, A, Wolf-Watz, M, Sauer, U.H, Sauer-Eriksson, A.E. | | Deposit date: | 2021-06-18 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Insights into the evolution of enzymatic specificity and catalysis: From Asgard archaea to human adenylate kinases.
Sci Adv, 8, 2022
|
|
7OWJ
 
 | | Odinarchaeota Adenylate kinase (OdinAK) in complex with GTP | | Descriptor: | Adenylate kinase, GUANOSINE-5'-TRIPHOSPHATE | | Authors: | Aberg-Zingmark, E, Grundstrom, C, Verma, A, Wolf-Watz, M, Sauer, U.H, Sauer-Eriksson, A.E. | | Deposit date: | 2021-06-18 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights into the evolution of enzymatic specificity and catalysis: From Asgard archaea to human adenylate kinases.
Sci Adv, 8, 2022
|
|
1QJF
 
 | | ISOPENICILLIN N SYNTHASE FROM ASPERGILLUS NIDULANS (Monocyclic Sulfoxide - Fe COMPLEX) | | Descriptor: | 1-[(1S)-CARBOXY-2-(METHYLSULFINYL)ETHYL]-(3R)-[(5S)-5-AMINO-5-CARBOXYPENTANAMIDO]-(4R)-SULFANYLAZETIDIN-2-ONE, FE (II) ION, ISOPENICILLIN N SYNTHASE, ... | | Authors: | Rutledge, P.J, Clifton, I.J, Burzlaff, N.I, Roach, P.L, Adlington, R.M, Baldwin, J.E. | | Deposit date: | 1999-06-23 | | Release date: | 2000-06-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Reaction Cycle of Isopenicillin N Synthase Observed by X-Ray Diffraction
Nature, 401, 1999
|
|
1QJE
 
 | | Isopenicillin N synthase from Aspergillus nidulans (IP1 - Fe complex) | | Descriptor: | FE (II) ION, ISOPENICILLIN N, ISOPENICILLIN N SYNTHASE, ... | | Authors: | Burzlaff, N.I, Clifton, I.J, Rutledge, P.J, Roach, P.L, Adlington, R.M, Baldwin, J.E. | | Deposit date: | 1999-06-23 | | Release date: | 2000-06-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The Reaction Cycle of Isopenicillin N Synthase Observed by X-Ray Diffraction
Nature, 401, 1999
|
|
3M4Z
 
 | | Crystal Structure of B. subtilis ferrochelatase with Cobalt bound at the active site | | Descriptor: | CHLORIDE ION, COBALT (II) ION, Ferrochelatase, ... | | Authors: | Soderberg, C.A.G, Hansson, M.D, Sreekanth, R, Al-Karadaghi, S, Hansson, M. | | Deposit date: | 2010-03-12 | | Release date: | 2010-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Bacterial ferrochelatase goes human: Tyr13 determines the apparent metal specificity of Bacillus subtilis ferrochelatase
To be Published
|
|
4BCX
 
 | | gamma 2 adaptin EAR domain crystal structure | | Descriptor: | 1,3-PROPANDIOL, AP-1 COMPLEX SUBUNIT GAMMA-LIKE 2, IMIDAZOLE | | Authors: | Juergens, M.C, Voros, J, Rautureau, G, Shepherd, D, Pye, V.E, Muldoon, J, Johnson, C.M, Ashcroft, A, Freund, S.M.V, Ferguson, N. | | Deposit date: | 2012-10-03 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Hepatitis B Virus Pres1 Domain Hijacks Host Trafficking Proteins by Motif Mimicry.
Nat.Chem.Biol., 9, 2013
|
|
1GAI
 
 | | GLUCOAMYLASE-471 COMPLEXED WITH D-GLUCO-DIHYDROACARBOSE | | Descriptor: | 4,6-dideoxy-4-{[(1S,2S,3S,4R,5R)-2,3,4-trihydroxy-5-(hydroxymethyl)cyclohexyl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, GLUCOAMYLASE-471, alpha-D-mannopyranose, ... | | Authors: | Aleshin, A.E, Stoffer, B, Firsov, L.M, Svensson, B, Honzatko, R.B. | | Deposit date: | 1996-03-06 | | Release date: | 1996-08-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystallographic complexes of glucoamylase with maltooligosaccharide analogs: relationship of stereochemical distortions at the nonreducing end to the catalytic mechanism.
Biochemistry, 35, 1996
|
|
1GAH
 
 | | GLUCOAMYLASE-471 COMPLEXED WITH ACARBOSE | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, GLUCOAMYLASE-471, alpha-D-mannopyranose, ... | | Authors: | Aleshin, A.E, Stoffer, B, Firsov, L.M, Svensson, B, Honzatko, R.B. | | Deposit date: | 1996-03-06 | | Release date: | 1996-08-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic complexes of glucoamylase with maltooligosaccharide analogs: relationship of stereochemical distortions at the nonreducing end to the catalytic mechanism.
Biochemistry, 35, 1996
|
|
