6E5L
 
 | | Crystal structure of human cellular retinol binding protein 1 in complex with abnormal-cannabidiol (abn-CBD) | | Descriptor: | (1'R,2'R)-5'-methyl-6-pentyl-2'-(prop-1-en-2-yl)-1',2',3',4'-tetrahydro[1,1'-biphenyl]-2,4-diol, Retinol-binding protein 1 | | Authors: | Silvaroli, J.A, Banerjee, S, Kiser, P.D, Golczak, M. | | Deposit date: | 2018-07-20 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Abnormal Cannabidiol Modulates Vitamin A Metabolism by Acting as a Competitive Inhibitor of CRBP1.
Acs Chem.Biol., 14, 2019
|
|
7TTZ
 
 | | Heterodimeric IgA Fc in complex with Staphylococcus aureus protein SSL7 | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Boulanger, M.J, Verstraete, M, Heinkel, F, Escobar, E, Dixit, S, Von Kreudenstein, T.S. | | Deposit date: | 2022-02-02 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Engineering a pure and stable heterodimeric IgA for the development of multispecific therapeutics.
Mabs, 14, 2022
|
|
6E5W
 
 | | Crystal structure of human cellular retinol binding protein 3 in complex with abnormal-cannabidiol (abn-CBD) | | Descriptor: | (1'R,2'R)-5'-methyl-6-pentyl-2'-(prop-1-en-2-yl)-1',2',3',4'-tetrahydro[1,1'-biphenyl]-2,4-diol, GLYCEROL, Retinol-binding protein 5 | | Authors: | Silvaroli, J.A, Banerjee, S, Kiser, P.D, Golczak, M. | | Deposit date: | 2018-07-23 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Abnormal Cannabidiol Modulates Vitamin A Metabolism by Acting as a Competitive Inhibitor of CRBP1.
Acs Chem.Biol., 14, 2019
|
|
8QZ8
 
 | |
7TXY
 
 | | Crystal structure of the 2-Aminophenol 1,6-dioxygenase from the ARO bacterial microcompartment of Micromonospora rosaria | | Descriptor: | 2-amino-5-chlorophenol 1,6-dioxygenase subunit alpha, 2-aminophenol 1,6-dioxygenase subunit beta, FE (II) ION | | Authors: | Sutter, M, Doron, L, Kerfeld, C.A. | | Deposit date: | 2022-02-10 | | Release date: | 2023-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Characterization of a novel aromatic substrate-processing microcompartment in Actinobacteria.
Mbio, 14, 2023
|
|
8QOH
 
 | | Crystal structure of the kinetoplastid kinetochore protein KKT14 C-terminal domain from Apiculatamorpha spiralis | | Descriptor: | kinetochore protein KKT14 | | Authors: | Carter, W, Ballmer, D, Ishii, M, Ludzia, P, Akiyoshi, B. | | Deposit date: | 2023-09-28 | | Release date: | 2024-01-17 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Kinetoplastid kinetochore proteins KKT14-KKT15 are divergent Bub1/BubR1-Bub3 proteins.
Open Biology, 14, 2024
|
|
7TVI
 
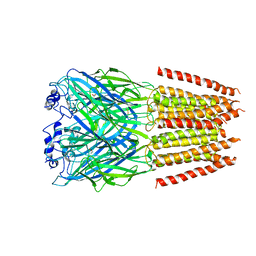 | | Alpha1/BetaB Heteromeric Glycine Receptor in Glycine-Bound State | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCINE, Glycine receptor beta subunit 2, ... | | Authors: | Gibbs, E, Chakrapani, S, Kumar, A. | | Deposit date: | 2022-02-04 | | Release date: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Conformational transitions and allosteric modulation in a heteromeric glycine receptor
Nat Commun, 14, 2023
|
|
7TU9
 
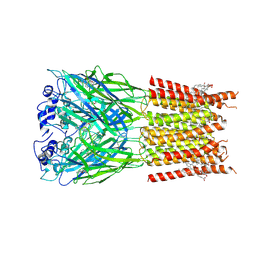 | | Alpha1/BetaB Heteromeric Glycine Receptor in Strychnine-Bound State | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Glycine receptor beta subunit 2, ... | | Authors: | Gibbs, E, Kumar, A, Chakrapani, S. | | Deposit date: | 2022-02-02 | | Release date: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Conformational transitions and allosteric modulation in a heteromeric glycine receptor
Nat Commun, 14, 2023
|
|
7TXF
 
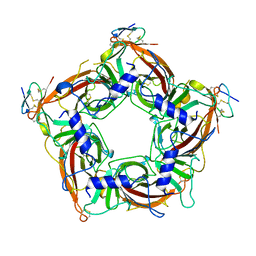 | |
7TQ1
 
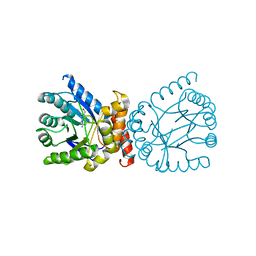 | | Crystal structure of adaptive laboratory evolved sulfonamide-resistant Dihydropteroate Synthase (DHPS) from Escherichia coli in complex with 6-hydroxymethylpterin | | Descriptor: | 6-HYDROXYMETHYLPTERIN, Dihydropteroate synthase | | Authors: | Stogios, P.J, Skarina, T, Tan, K, Venkatesan, M, Fruci, M, Joachimiak, A, Savchenko, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-26 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Molecular mechanism of plasmid-borne resistance to sulfonamide antibiotics.
Nat Commun, 14, 2023
|
|
8QZI
 
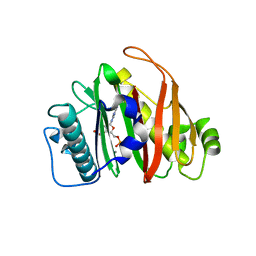 | | Crystal structure of PptT-ACP from Mycobacterium tuberculosis | | Descriptor: | 4'-phosphopantetheinyl transferase PptT, COENZYME A, ISOPROPYL ALCOHOL, ... | | Authors: | Gavalda, S, Mourey, L, Pedelacq, J.D. | | Deposit date: | 2023-10-27 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Catalytic Cycle of Type II 4'-Phosphopantetheinyl Transferases
Acs Catalysis, 14, 2024
|
|
8QZH
 
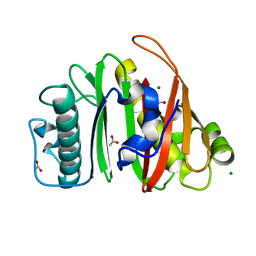 | | Crystal structure of apo-PptT from Mycobacterium tuberculosis | | Descriptor: | 4'-phosphopantetheinyl transferase PptT, ACETATE ION, MAGNESIUM ION | | Authors: | Gavalda, S, Carivenc, C, Mourey, L, Pedelacq, J.D. | | Deposit date: | 2023-10-27 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Catalytic Cycle of Type II 4'-Phosphopantetheinyl Transferases
Acs Catalysis, 14, 2024
|
|
6E6K
 
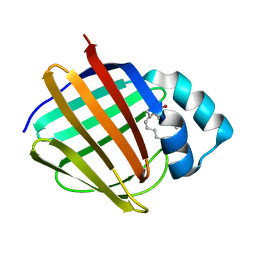 | | Crystal structure of human cellular retinol-binding protein 4 in complex with abnormal-cannabidiol (abn-CBD) | | Descriptor: | (1'R,2'R)-5'-methyl-6-pentyl-2'-(prop-1-en-2-yl)-1',2',3',4'-tetrahydro[1,1'-biphenyl]-2,4-diol, Retinoid-binding protein 7 | | Authors: | Silvaroli, J.A, Banerjee, S, Kiser, P.D, Golczak, M. | | Deposit date: | 2018-07-25 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Abnormal Cannabidiol Modulates Vitamin A Metabolism by Acting as a Competitive Inhibitor of CRBP1.
Acs Chem.Biol., 14, 2019
|
|
8QDC
 
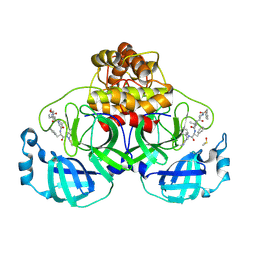 | | Crystal structure of SARS-CoV-2 main protease (MPro) in complex with the covalent inhibitor GUE-3642 (compound 1 in publication) | | Descriptor: | (phenylmethyl) ~{N}-[(2~{S})-1-[[(2~{S})-1-[[(2~{S})-1-[[iminomethyl-(phenylmethyl)amino]-methyl-amino]-1-oxidanylidene-3-phenyl-propan-2-yl]amino]-3,3-dimethyl-1-oxidanylidene-butan-2-yl]amino]-1-oxidanylidene-butan-2-yl]carbamate, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | Strater, N, Claff, T, Sylvester, K, Mueller, C.E, Guetschow, M. | | Deposit date: | 2023-08-28 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Development of an active-site titrant for SARS-CoV-2 main protease as an indispensable tool for evaluating enzyme kinetics.
Acta Pharm Sin B, 14, 2024
|
|
6ELN
 
 | | Estimation of relative drug-target residence times by random acceleration molecular dynamics simulation | | Descriptor: | 4-[4-(4-methoxyphenyl)-5-methyl-1H-pyrazol-3-yl]benzene-1,3-diol, Heat shock protein HSP 90-alpha, SULFATE ION | | Authors: | Musil, D, Lehmann, M, Eggenweiler, H.-M. | | Deposit date: | 2017-09-29 | | Release date: | 2018-05-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Estimation of Drug-Target Residence Times by tau-Random Acceleration Molecular Dynamics Simulations.
J Chem Theory Comput, 14, 2018
|
|
6ELO
 
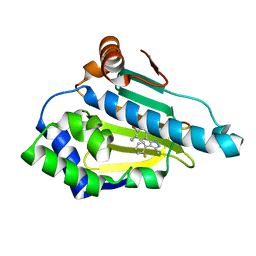 | |
7MRJ
 
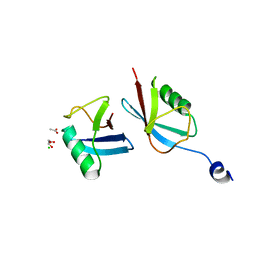 | | Crystal structure of a novel ubiquitin-like TINCR | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Forouhar, F, Morgado-Palacin, L, Brown, J.A, Martinez, T, Pedrero, J.M.G, Reglero, C, Chaudhry, I, Vaughan, J, Rodriguez-Perales, S, Allonca, E, Granda-Diaz, R, Quinn, S.A, Fernandez, A.F, Fraga, M.F, Kim, A.L, Santos-Juanes, J, Owens, D.M, Rodrigo, J.P, Saghatelian, A, Ferrando, A.A. | | Deposit date: | 2021-05-07 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | The TINCR ubiquitin-like microprotein is a tumor suppressor in squamous cell carcinoma.
Nat Commun, 14, 2023
|
|
8SBB
 
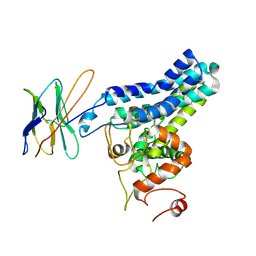 | | Cryo-EM structure of FtAlkB | | Descriptor: | Alkane 1-monooxygenase, DODECANE, FE (III) ION, ... | | Authors: | Zhang, J, Feng, L. | | Deposit date: | 2023-04-03 | | Release date: | 2023-04-26 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Structure and mechanism of the alkane-oxidizing enzyme AlkB.
Nat Commun, 14, 2023
|
|
8SCD
 
 | | Crystal structure of sulfonamide resistance enzyme Sul3 in complex with reaction intermediate | | Descriptor: | 2-amino-6-methylidene-6,7-dihydropteridin-4(3H)-one, 4-AMINOBENZOIC ACID, CHLORIDE ION, ... | | Authors: | Stogios, P.J, Venkatesan, M, Michalska, K, Mesa, N, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2023-04-05 | | Release date: | 2023-05-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Molecular mechanism of plasmid-borne resistance to sulfonamide antibiotics.
Nat Commun, 14, 2023
|
|
8SI6
 
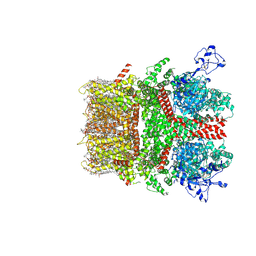 | | Cryo-EM structure of TRPM7 in MSP2N2 nanodisc in complex with agonist naltriben in closed state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, (4bS,8R,8aS,14bR)-7-(cyclopropylmethyl)-5,6,7,8,9,14b-hexahydro-8aH-4,8-methanobis[1]benzofuro[3,2-e:2',3'-g]isoquinoline-1,8a-diol, 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, ... | | Authors: | Nadezhdin, K.D, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2023-04-14 | | Release date: | 2023-05-17 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (2.44 Å) | | Cite: | Structural mechanisms of TRPM7 activation and inhibition.
Nat Commun, 14, 2023
|
|
8SI5
 
 | | Cryo-EM structure of TRPM7 in MSP2N2 nanodisc in complex with agonist naltriben in open state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, (4bS,8R,8aS,14bR)-7-(cyclopropylmethyl)-5,6,7,8,9,14b-hexahydro-8aH-4,8-methanobis[1]benzofuro[3,2-e:2',3'-g]isoquinoline-1,8a-diol, 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, ... | | Authors: | Nadezhdin, K.D, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2023-04-14 | | Release date: | 2023-05-17 | | Method: | ELECTRON MICROSCOPY (2.17 Å) | | Cite: | Structural mechanisms of TRPM7 activation and inhibition.
Nat Commun, 14, 2023
|
|
8SI4
 
 | | Cryo-EM structure of TRPM7 N1098Q mutant in GDN detergent in open state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, CALCIUM ION, ... | | Authors: | Nadezhdin, K.D, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2023-04-14 | | Release date: | 2023-05-17 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (2.46 Å) | | Cite: | Structural mechanisms of TRPM7 activation and inhibition.
Nat Commun, 14, 2023
|
|
8SI7
 
 | | Cryo-EM structure of TRPM7 in GDN detergent in complex with inhibitor VER155008 in closed state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, 4-[[(2R,3S,4R,5R)-5-[6-amino-8-[(3,4-dichlorophenyl)methylamino]purin-9-yl]-3,4-dihydroxy-oxolan-2-yl]methoxymethyl]benzonitrile, ... | | Authors: | Nadezhdin, K.D, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2023-04-14 | | Release date: | 2023-05-17 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Structural mechanisms of TRPM7 activation and inhibition.
Nat Commun, 14, 2023
|
|
8SIB
 
 | |
8SIA
 
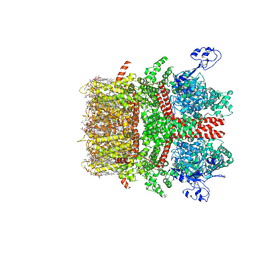 | | Cryo-EM structure of TRPM7 N1098Q mutant in GDN detergent in complex with inhibitor NS8593 in closed state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, CALCIUM ION, ... | | Authors: | Nadezhdin, K.D, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2023-04-14 | | Release date: | 2023-05-17 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Structural mechanisms of TRPM7 activation and inhibition.
Nat Commun, 14, 2023
|
|
