4M3K
 
 | | Structure of a single domain camelid antibody fragment cAb-H7S in complex with the BlaP beta-lactamase from Bacillus licheniformis | | Descriptor: | Beta-lactamase, CHLORIDE ION, Camelid heavy-chain antibody variable fragment cAb-H7S | | Authors: | Pain, C, Kerff, F, Herman, R, Sauvage, E, Preumont, S, Charlier, P, Dumoulin, M. | | Deposit date: | 2013-08-06 | | Release date: | 2014-08-06 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Probing the mechanism of aggregation of polyQ model proteins with camelid heavy-chain antibody fragments
To be Published
|
|
4JXG
 
 | | Crystal Structure of AmpC beta-lactamase from E. coli in Complex with Oxacillin | | Descriptor: | (2R,4S)-5,5-dimethyl-2-[(1R)-1-{[(5-methyl-3-phenyl-1,2-oxazol-4-yl)carbonyl]amino}-2-oxoethyl]-1,3-thiazolidine-4-carb oxylic acid, Beta-lactamase, PHOSPHATE ION, ... | | Authors: | Wallar, B.J, Powers, R.A, Docter, B.E. | | Deposit date: | 2013-03-28 | | Release date: | 2014-10-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Complexed structures of AmpC beta-lactamase
To be Published
|
|
1FCN
 
 | |
4KEN
 
 | | Crystal Structure of AmpC beta-lactamase N152G Mutant in Complex with Cefoxitin | | Descriptor: | (2R)-2-{(1S)-1-methoxy-2-oxo-1-[(thiophen-2-ylacetyl)amino]ethyl}-5-methylidene-5,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Docter, B.E, Baggett, V.L, Powers, R.A, Wallar, B.J. | | Deposit date: | 2013-04-25 | | Release date: | 2014-10-29 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Complexed structures of AmpC beta-lactamase
To be Published
|
|
4KG5
 
 | | Crystal Structure of AmpC beta-lactamase N152G Mutant in Complex with Cefotaxime | | Descriptor: | Beta-lactamase, CEFOTAXIME, C3' cleaved, ... | | Authors: | Docter, B.E, Baggett, V.L, Powers, R.A, Wallar, B.J. | | Deposit date: | 2013-04-28 | | Release date: | 2014-10-29 | | Last modified: | 2018-09-19 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Complexed structures of AmpC beta-lactamase
To be Published
|
|
1O07
 
 | | Crystal Structure of the complex between Q120L/Y150E mutant of AmpC and a beta-lactam inhibitor (MXG) | | Descriptor: | 2-(1-{2-[4-(2-ACETYLAMINO-PROPIONYLAMINO)-4-CARBOXY-BUTYRYLAMINO]-6-AMINO-HEXANOYLAMINO}-2-OXO-ETHYL)-5-METHYLENE-5,6-DIHYDRO-2H-[1,3]THIAZINE-4-CARBOXYLIC ACID, Beta-lactamase, POTASSIUM ION | | Authors: | Meroueh, S.O, Minasov, G, Lee, W, Shoichet, B.K, Mobashery, S. | | Deposit date: | 2003-02-20 | | Release date: | 2003-08-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structural Aspects for Evolution of beta-Lactamases from Penicillin-Binding Proteins
J.Am.Chem.Soc., 125, 2003
|
|
4QD4
 
 | | Structure of ADC-68, a Novel Carbapenem-Hydrolyzing Class C Extended-Spectrum -Lactamase from Acinetobacter baumannii | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-lactamase ADC-68, CITRIC ACID | | Authors: | Hong, M.K, Kang, L.W. | | Deposit date: | 2014-05-13 | | Release date: | 2015-01-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of ADC-68, a novel carbapenem-hydrolyzing class C extended-spectrum beta-lactamase isolated from Acinetobacter baumannii
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4KG2
 
 | | Crystal Structure of AmpC beta-lactamase from E. coli in Complex with Cefotaxime | | Descriptor: | Beta-lactamase, CEFOTAXIME, C3' cleaved, ... | | Authors: | Docter, B.E, Baggett, V.L, Powers, R.A, Wallar, B.J. | | Deposit date: | 2013-04-28 | | Release date: | 2014-10-29 | | Last modified: | 2018-09-19 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Complexed structures of AmpC beta-lactamase
To be Published
|
|
4JXS
 
 | |
1GA9
 
 | | CRYSTAL STRUCTURE OF AMPC BETA-LACTAMASE FROM E. COLI COMPLEXED WITH NON-BETA-LACTAMASE INHIBITOR (2, 3-(4-BENZENESULFONYL-THIOPHENE-2-SULFONYLAMINO)-PHENYLBORONIC ACID) | | Descriptor: | 3-(4-BENZENESULFONYL-THIOPHENE-2-SULFONYLAMINO)-PHENYLBORONIC ACID, BETA-LACTAMASE, PHOSPHATE ION, ... | | Authors: | Tondi, D, Powers, R.A, Caselli, E, Negri, M.C, Blazquez, J, Costi, M.P, Shoichet, B.K. | | Deposit date: | 2000-11-29 | | Release date: | 2001-07-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based design and in-parallel synthesis of inhibitors of AmpC beta-lactamase.
Chem.Biol., 8, 2001
|
|
1KE3
 
 | |
1IEM
 
 | | Crystal Structure of AmpC beta-lactamase from E. coli in Complex with a Boronic Acid Inhibitor (1, CefB4) | | Descriptor: | PHOSPHATE ION, PINACOL[[2-AMINO-ALPHA-(1-CARBOXY-1-METHYLETHOXYIMINO)-4-THIAZOLEACETYL]AMINO]METHANEBORONATE, beta-lactamase | | Authors: | Powers, R.A, Caselli, E, Focia, P.J, Prati, F, Shoichet, B.K. | | Deposit date: | 2001-04-10 | | Release date: | 2001-08-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of ceftazidime and its transition-state analogue in complex with AmpC beta-lactamase: implications for resistance mutations and inhibitor design.
Biochemistry, 40, 2001
|
|
1IKG
 
 | | MICHAELIS COMPLEX OF STREPTOMYCES R61 DD-PEPTIDASE WITH A SPECIFIC PEPTIDOGLYCAN SUBSTRATE FRAGMENT | | Descriptor: | D-ALANYL-D-ALANINE CARBOXYPEPTIDASE, GLYCYL-L-ALPHA-AMINO-EPSILON-PIMELYL-D-ALANYL-D-ALANINE | | Authors: | Mcdonough, M.A, Anderson, J.W, Silvaggi, N.R, Pratt, R.F, Knox, J.R, Kelly, J.A. | | Deposit date: | 2001-05-03 | | Release date: | 2002-09-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of two kinetic intermediates reveal species specificity of penicillin-binding proteins.
J.Mol.Biol., 322, 2002
|
|
1IEL
 
 | | Crystal Structure of AmpC beta-lactamase from E. coli in Complex with Ceftazidime | | Descriptor: | ACYLATED CEFTAZIDIME, PHOSPHATE ION, beta-lactamase | | Authors: | Powers, R.A, Caselli, E, Focia, P.J, Prati, F, Shoichet, B.K. | | Deposit date: | 2001-04-10 | | Release date: | 2001-08-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of ceftazidime and its transition-state analogue in complex with AmpC beta-lactamase: implications for resistance mutations and inhibitor design.
Biochemistry, 40, 2001
|
|
1GCE
 
 | | STRUCTURE OF THE BETA-LACTAMASE OF ENTEROBACTER CLOACAE GC1 | | Descriptor: | BETA-LACTAMASE | | Authors: | Crichlow, G.V, Kuzin, A.P, Nukaga, M, Sawai, T, Knox, J.R. | | Deposit date: | 1999-05-17 | | Release date: | 1999-08-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the extended-spectrum class C beta-lactamase of Enterobacter cloacae GC1, a natural mutant with a tandem tripeptide insertion.
Biochemistry, 38, 1999
|
|
4OLG
 
 | | Crystal structure of AmpC beta-lactamase in complex with covalently bound N-formyl 7-aminocephalosporanic acid | | Descriptor: | (2R,5Z)-5-[(acetyloxy)methylidene]-2-[(1R)-1-(formylamino)-2-oxoethyl]-5,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Shoichet, B.K, Barelier, S. | | Deposit date: | 2014-01-23 | | Release date: | 2014-05-28 | | Last modified: | 2014-06-18 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Substrate deconstruction and the nonadditivity of enzyme recognition.
J.Am.Chem.Soc., 136, 2014
|
|
3GQZ
 
 | |
3GTC
 
 | | AmpC beta-lactamase in complex with Fragment-based Inhibitor | | Descriptor: | (1R,2S)-2-(5-thioxo-4,5-dihydro-1H-1,2,4-triazol-3-yl)cyclohexanecarboxylic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Teotico, D.T, Shoichet, B.K. | | Deposit date: | 2009-03-27 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Docking for fragment inhibitors of AmpC beta-lactamase
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
6K8X
 
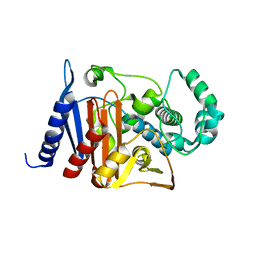 | | Crystal structure of a class C beta lactamase | | Descriptor: | Beta-lactamase | | Authors: | Bae, D.W, Jung, Y.E, An, Y.J, Na, J.H, Cha, S.S. | | Deposit date: | 2019-06-13 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural Insights into Catalytic Relevances of Substrate Poses in ACC-1.
Antimicrob.Agents Chemother., 63, 2019
|
|
6KA5
 
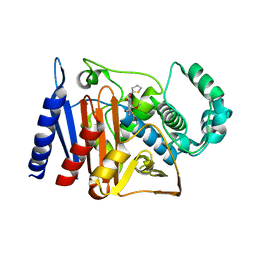 | | Crystal structure of a class C beta-lactamase in complex with cefoxitin | | Descriptor: | (2R)-2-{(1S)-1-methoxy-2-oxo-1-[(thiophen-2-ylacetyl)amino]ethyl}-5-methylidene-5,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Beta-lactamase | | Authors: | Bae, D.W, Jung, Y.E, An, Y.J, Na, J.H, Cha, S.S. | | Deposit date: | 2019-06-20 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural Insights into Catalytic Relevances of Substrate Poses in ACC-1.
Antimicrob.Agents Chemother., 63, 2019
|
|
6KJE
 
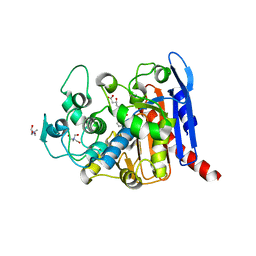 | | lovastatin esterase PcEST in complex with monacolin J | | Descriptor: | (3R,5R)-3,5-dihydroxy-7-[(1S,2S,6R,8S,8aR)-8-hydroxy-2,6-dimethyl-1,2,6,7,8,8a-hexahydronaphthalen-1-yl]heptanoic acid, ACETATE ION, Pc15g00720 protein, ... | | Authors: | Liang, Y.J, Lu, X.F. | | Deposit date: | 2019-07-22 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.484 Å) | | Cite: | Structural insights into the catalytic mechanism of lovastatin hydrolase
J.Biol.Chem., 2019
|
|
4E3M
 
 | |
6KJR
 
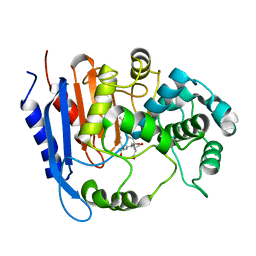 | | Functional and structural insights into the unusual oxyanion hole-like geometry in macrolactin acyltransferase selective for dicarboxylic acyl donors | | Descriptor: | 4-[[(3~{E},5~{Z},8~{S},9~{E},11~{E},14~{S},16~{R},17~{Z},19~{E},24~{R})-24-methyl-14,16-bis(oxidanyl)-2-oxidanylidene-1-oxacyclotetracosa-3,5,9,11,17,19-hexaen-8-yl]oxy]-4-oxidanylidene-butanoic acid, Putative beta-lactamase | | Authors: | Xiao, F, Dong, S, Feng, Y, Li, W. | | Deposit date: | 2019-07-23 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.361 Å) | | Cite: | Structural Basis of Specificity for Carboxyl-Terminated Acyl Donors in a Bacterial Acyltransferase.
J.Am.Chem.Soc., 142, 2020
|
|
4E3O
 
 | |
6KJP
 
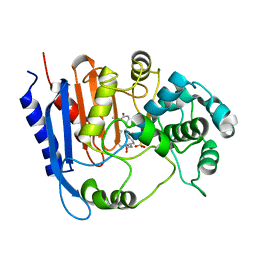 | | Functional and structural insights into the unusual oxyanion hole-like geometry in macrolactin acyltransferase selective for dicarboxylic acyl donors | | Descriptor: | (3~{Z},5~{E},8~{S},9~{E},11~{E},14~{S},16~{R},17~{Z},19~{E},24~{R})-24-methyl-8,14,16-tris(oxidanyl)-1-oxacyclotetracosa-3,5,9,11,17,19-hexaen-2-one, Putative beta-lactamase, SULFATE ION | | Authors: | Xiao, F, Dong, S, Feng, Y, Li, W. | | Deposit date: | 2019-07-23 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural Basis of Specificity for Carboxyl-Terminated Acyl Donors in a Bacterial Acyltransferase.
J.Am.Chem.Soc., 142, 2020
|
|
