8YYR
 
 | | Structure of the HitB T293G mutant | | Descriptor: | Putative ATP-dependent b-aminoacyl-ACP synthetase, [(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl ~{N}-[(3~{S})-3-azanyl-3-(2-bromophenyl)propanoyl]sulfamate | | Authors: | Wang, D, Miyanaga, A, Chisuga, T, Kudo, F, Eguchi, T. | | Deposit date: | 2024-04-04 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Engineering the Substrate Specificity of (S)-beta-Phenylalanine Adenylation Enzyme HitB.
Chembiochem, 2024
|
|
8UTX
 
 | | Solution structure of a 12-mer peptide bearing a bicyclic Asx motif mimic (BAMM) as a synthetic N-cap | | Descriptor: | 1,3,5-tris(bromomethyl)benzene, TRP-CYS-ASP-ALA-ALA-CYS-CYS-ALA-ALA-ALA-LYS-ALA-NH2 peptide | | Authors: | Mi, T.X, Burgess, K. | | Deposit date: | 2023-10-31 | | Release date: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | Bioinformatics leading to conveniently accessible, helix enforcing, bicyclic ASX motif mimics (BAMMs).
Nat Commun, 15, 2024
|
|
9BKT
 
 | |
8WUT
 
 | | SpCas9-MMLV RT-pegRNA-target DNA complex (initiation) | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, DNA (5'-D(*TP*GP*AP*TP*GP*GP*CP*AP*GP*AP*GP*TP*AP*CP*TP*AP*G)-3'), DNA (51-MER), ... | | Authors: | Shuto, Y, Nakagawa, R, Hoki, M, Omura, S.N, Hirano, H, Itoh, Y, Nureki, O. | | Deposit date: | 2023-10-21 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for pegRNA-guided reverse transcription by a prime editor.
Nature, 2024
|
|
8XAV
 
 | |
8YRP
 
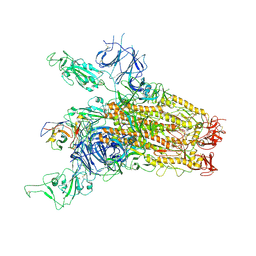 | | SARS-CoV-2 Delta Spike in complex with JM-1A | | Descriptor: | JM-1A Heavy Chain, JM-1A Light Chain, Spike glycoprotein | | Authors: | Nguyen, V.H.T, Chen, X. | | Deposit date: | 2024-03-21 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | The presence of broadly neutralizing anti-SARS-CoV-2 RBD antibodies elicited by primary series and booster dose of COVID-19 vaccine
Plos Pathog., 2024
|
|
8YUU
 
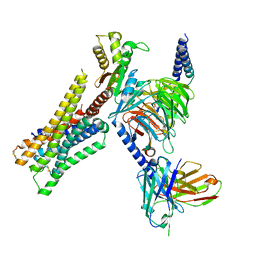 | | Cryo-EM structure of the histamine-bound H3R-Gi complex | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Shen, Q, Tang, X, Wen, X, Cheng, S, Xiao, P, Zang, S, Shen, D, Jiang, L, Zheng, Y, Zhang, H, Xu, H, Mao, C, Zhang, M, Hu, W, Sun, J, Chen, Z, Zhang, Y. | | Deposit date: | 2024-03-27 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Molecular Determinant Underlying Selective Coupling of Primary G-Protein by Class A GPCRs.
Adv Sci, 2024
|
|
8YXR
 
 | |
8ZEY
 
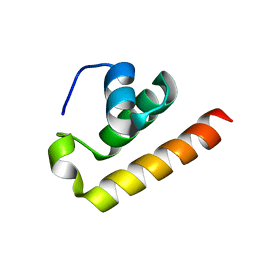 | | Anti-CRISPR type I subtype E3;AcrIE3 | | Descriptor: | AcrIE3 | | Authors: | Kim, D.Y, Park, H.H. | | Deposit date: | 2024-05-07 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.734 Å) | | Cite: | Novel structure of the anti-CRISPR protein AcrIE3 and its implication on the CRISPR-Cas inhibition.
Biochem.Biophys.Res.Commun., 722, 2024
|
|
8ZMU
 
 | |
8ZN3
 
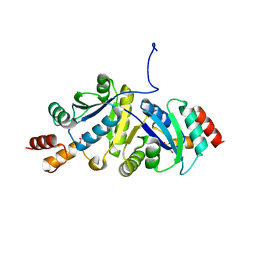 | | Structure of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter sp. with the expression tag bound in the substrate binding site of a neighbouring molecule at 2.41 A resolution. | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, PHOSPHONOACETIC ACID, ... | | Authors: | Ahmad, N, Sharma, P, Bhushan, A, Sharma, S, Singh, T.P. | | Deposit date: | 2024-05-25 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structure of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter sp. with the expression tag bound in the substrate binding site of a neighbouring molecule at 2.41 A resolution.
To Be Published
|
|
9B58
 
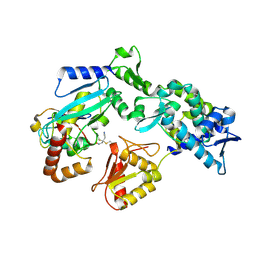 | |
9B59
 
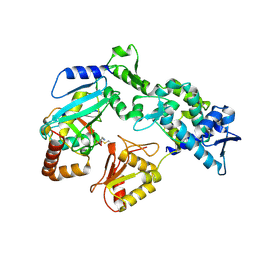 | |
8XBS
 
 | | C. elegans apo-SID1 structure | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gong, D.S. | | Deposit date: | 2023-12-07 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.21 Å) | | Cite: | Structural basis for double-stranded RNA recognition by SID1.
Nucleic Acids Res., 2024
|
|
9B5N
 
 | |
8Y2H
 
 | |
9B5W
 
 | |
9BKF
 
 | |
8YXP
 
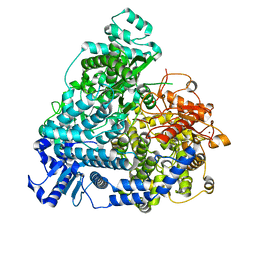 | | Structure of mumps virus L protein (state2) | | Descriptor: | RNA-directed RNA polymerase L, ZINC ION | | Authors: | Li, T.H, Shen, Q.T. | | Deposit date: | 2024-04-02 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structures of the mumps virus polymerase complex via cryo-electron microscopy.
Nat Commun, 15, 2024
|
|
9BKP
 
 | |
9B5B
 
 | |
9F3E
 
 | |
8X8S
 
 | |
9B5J
 
 | |
9BB5
 
 | |
