3LW0
 
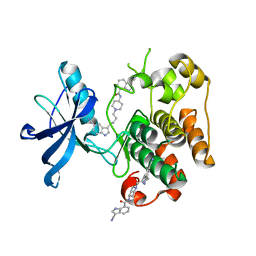 | | IGF-1RK in complex with ligand MSC1609119A-1 | | Descriptor: | 3-cyano-N-{1-[4-(5-cyano-1H-indol-3-yl)butyl]piperidin-4-yl}-1H-indole-7-carboxamide, GLYCEROL, Insulin-like growth factor 1 receptor | | Authors: | Graedler, U, Heinrich, T, Boettcher, H, Blaukat, A, Shutes, A, Askew, B. | | Deposit date: | 2010-02-23 | | Release date: | 2010-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Allosteric IGF-1R Inhibitors.
Acs Med.Chem.Lett., 1, 2010
|
|
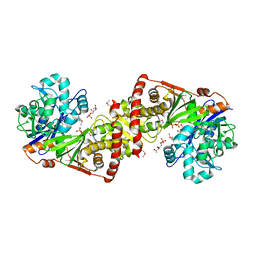
3LL3
 
 | |
3LT8
 
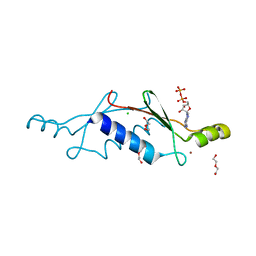 | | A non-biological ATP binding protein with a single point mutation (D65V), that contributes to optimized folding and ligand binding, crystallized in the presence of 100 mM ATP. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP BINDING PROTEIN-D65V, CHLORIDE ION, ... | | Authors: | Simmons, C.R, Magee, C.L, Allen, J.P, Chaput, J.C. | | Deposit date: | 2010-02-15 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Three-dimensional structures reveal multiple ADP/ATP binding modes for a synthetic class of artificial proteins.
Biochemistry, 49, 2010
|
|
3LT9
 
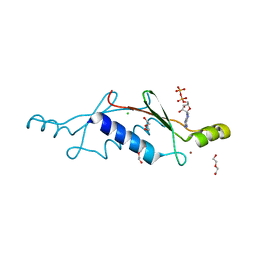 | | A non-biological ATP binding protein with a single point mutation (D65V), that contributes to optimized folding and ligand binding | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP BINDING PROTEIN-D65V, CHLORIDE ION, ... | | Authors: | Simmons, C.R, Magee, C.L, Allen, J.P, Chaput, J.C. | | Deposit date: | 2010-02-15 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Three-dimensional structures reveal multiple ADP/ATP binding modes for a synthetic class of artificial proteins.
Biochemistry, 49, 2010
|
|
4DT2
 
 | | Crystal structure of red kidney bean purple acid phosphatase in complex with Maybridge fragment CC27209 | | Descriptor: | (2,2-dimethyl-2,3-dihydro-1-benzofuran-7-yl)methanol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Feder, D, Hussein, W.M, Clayton, D.J, Kan, M, Schenk, G, McGeary, R.P, Guddat, L.W. | | Deposit date: | 2012-02-20 | | Release date: | 2012-09-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification of purple acid phosphatase inhibitors by fragment-based screening: promising new leads for osteoporosis therapeutics.
Chem.Biol.Drug Des., 80, 2012
|
|
6VG3
 
 | |
4YVC
 
 | | ROCK 1 bound to thiazole inhibitor | | Descriptor: | 2-fluoro-N-[4-(pyridin-4-yl)-1,3-thiazol-2-yl]benzamide, Rho-associated protein kinase 1 | | Authors: | Jacobs, M.D. | | Deposit date: | 2015-03-19 | | Release date: | 2015-06-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Design, Synthesis, and Structure-Activity Relationships of Pyridine-Based Rho Kinase (ROCK) Inhibitors.
J.Med.Chem., 58, 2015
|
|
3LTA
 
 | | Crystal structure of a non-biological ATP binding protein with a TYR-PHE mutation within the ligand binding domain | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP BINDING PROTEIN-DX, CHLORIDE ION, ... | | Authors: | Simmons, C.R, Magee, C.L, Allen, J.P, Chaput, J.C. | | Deposit date: | 2010-02-15 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Three-dimensional structures reveal multiple ADP/ATP binding modes for a synthetic class of artificial proteins.
Biochemistry, 49, 2010
|
|
5QLV
 
 | |
5QMA
 
 | |
5QMF
 
 | |
5QMP
 
 | |
5QMV
 
 | |
5QN7
 
 | |
5QNC
 
 | |
5IIE
 
 | |
5QMI
 
 | |
5QN0
 
 | |
5QKF
 
 | |
5QKY
 
 | |
5QLA
 
 | |
5QLU
 
 | |
5QMB
 
 | |
5QMR
 
 | |
5QN6
 
 | |
