3GH0
 
 | | Replacement of Val3 in Human Thymidylate Synthase Affects Its Kinetic Properties and Intracellular Stability | | Descriptor: | SULFATE ION, Thymidylate synthase | | Authors: | Huang, X, Gibson, L.M, Bell, B.J, Lovelace, L.L, Lebioda, L. | | Deposit date: | 2009-03-02 | | Release date: | 2010-03-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Replacement of Val3 in human thymidylate synthase affects its kinetic properties and intracellular stability .
Biochemistry, 49, 2010
|
|
3GGM
 
 | | Crystal Structure of BT9727_2919 from Bacillus thuringiensis subsp. Northeast Structural Genomics Target BuR228B | | Descriptor: | uncharacterized protein BT9727_2919 | | Authors: | Seetharaman, J, Neely, H, Wang, H, Janjua, H, Foote, E.L, Xiao, R, Everett, J.K, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-02-28 | | Release date: | 2009-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of BT9727_2919 from Bacillus thuringiensis subsp. Northeast Structural Genomics Target BuR228B
To be Published
|
|
2WWR
 
 | | Crystal Structure of Human Glyoxylate Reductase Hydroxypyruvate Reductase | | Descriptor: | GLYOXYLATE REDUCTASE/HYDROXYPYRUVATE REDUCTASE, MAGNESIUM ION | | Authors: | Booth, M.P.S, Conners, R, Rumsby, G, Brady, R.L. | | Deposit date: | 2009-10-26 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Structural Basis of Substrate Specificity in Human Glyoxylate Reductase/Hydroxypyruvate Reductase.
J.Mol.Biol., 360, 2006
|
|
3KMH
 
 | |
5YWN
 
 | | SsCR_L211H-NADP+ | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Protein induced by osmotic stress | | Authors: | Shang, Y.P, Chen, Q, Yu, H.L, Xu, J.H. | | Deposit date: | 2017-11-29 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.039 Å) | | Cite: | Attenuated substrate inhibition of a haloketone reductase via structure-guided loop engineering.
J.Biotechnol., 308, 2020
|
|
6CT6
 
 | | Crystal structure of lactate dehydrogenase from Eimeria maxima with NADH and oxamate | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Lactate dehydrogenase, OXAMIC ACID, ... | | Authors: | Wirth, J.D, Xu, C, Theobald, D.L. | | Deposit date: | 2018-03-22 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.705 Å) | | Cite: | The Mechanistic, Structural, and Evolutionary Origin of Lactate Dehydrogenase Substrate Specificity in Apicomplexa
To Be Published
|
|
7XVH
 
 | | Crystal structure of AtHPPD-Y13287 complex | | Descriptor: | 1,5-dimethyl-3-(2-methylphenyl)-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-quinazoline-2,4-dione, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Lin, H.-Y, Dong, J, Yang, G.-F. | | Deposit date: | 2022-05-23 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Discovery of Subnanomolar Inhibitors of 4-Hydroxyphenylpyruvate Dioxygenase via Structure-Based Rational Design.
J.Agric.Food Chem., 71, 2023
|
|
2X0R
 
 | | R207S, R292S Mutant of Malate Dehydrogenase from the Halophilic Archeon Haloarcula marismortui (HoloForm) | | Descriptor: | CHLORIDE ION, MALATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Irimia, A, Ebel, C, Vellieux, F.M.D, Richard, S.B, Cosenza, L.W, Zaccai, G, Madern, D. | | Deposit date: | 2009-12-17 | | Release date: | 2009-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.915 Å) | | Cite: | The Oligomeric States of Haloarcula Marismortui Malate Dehydrogenase are Modulated by Solvent Components as Shown by Crystallographic and Biochemical Studies
J.Mol.Biol., 326, 2003
|
|
6CYA
 
 | | Rotavirus SA11 NSP2 S313A mutant | | Descriptor: | CHLORIDE ION, GLYCEROL, Non-structural protein 2 | | Authors: | Anish, R, Hu, L, Prasad, B.V.V. | | Deposit date: | 2018-04-05 | | Release date: | 2018-12-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Phosphorylation cascade regulates the formation and maturation of rotaviral replication factories.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2X2N
 
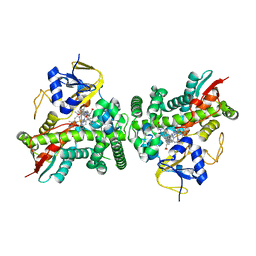 | | X-ray structure of cyp51 from trypanosoma brucei in complex with posaconazole in two different conformations | | Descriptor: | LANOSTEROL 14-ALPHA-DEMETHYLASE, POSACONAZOLE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Chen, C.-K, Leung, S.S.F, Guilbert, C, Jacobson, M, McKerrow, J.H, Podust, L.M. | | Deposit date: | 2010-01-14 | | Release date: | 2010-02-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Characterization of Cyp51 from Trypanosoma Cruzi and Trypanosoma Brucei Bound to the Antifungal Drugs Posaconazole and Fluconazole
Plos Negl Trop Dis, 4, 2010
|
|
4NCS
 
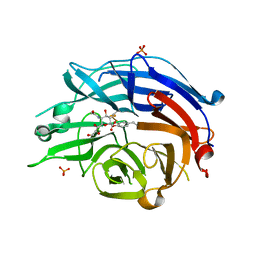 | | Human sialidase 2 in complex with 2,3-difluorosialic acid (covalent intermediate) | | Descriptor: | (2S,3S,4R,5R,6R)-5-acetamido-2,3-bis(fluoranyl)-4-oxidanyl-6-[(1S,2S)-1,2,3-tris(oxidanyl)propyl]oxane-2-carboxylic acid, PHOSPHATE ION, Sialidase-2 | | Authors: | Buchini, S, Gallat, F.-X, Greig, I.R, Kim, J.-H, Wakatsuki, S, Chavas, L.M.G, Withers, S.G. | | Deposit date: | 2013-10-25 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Tuning mechanism-based inactivators of neuraminidases: mechanistic and structural insights.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4ND2
 
 | |
7Y7A
 
 | | In situ double-PBS-PSII-PSI-LHCs megacomplex from Porphyridium purpureum. | | Descriptor: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, (2S)-2,3-dihydroxypropyl octadecanoate, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | You, X, Zhang, X, Cheng, J, Xiao, Y.N, Sun, S, Sui, S.F. | | Deposit date: | 2022-06-22 | | Release date: | 2023-02-08 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | In situ structure of the red algal phycobilisome-PSII-PSI-LHC megacomplex.
Nature, 616, 2023
|
|
4NDF
 
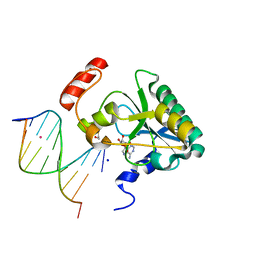 | | Human Aprataxin (Aptx) bound to RNA-DNA, AMP, and Zn - product complex | | Descriptor: | 5'-D(*GP*AP*AP*TP*CP*AP*TP*AP*AP*C)-3', 5'-R(P*G)-D(P*TP*TP*AP*TP*GP*AP*TP*TP*C)-3', ADENOSINE MONOPHOSPHATE, ... | | Authors: | Schellenberg, M.J, Tumbale, P.S, Williams, R.S. | | Deposit date: | 2013-10-26 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.944 Å) | | Cite: | Aprataxin resolves adenylated RNA-DNA junctions to maintain genome integrity.
Nature, 506, 2013
|
|
4NDG
 
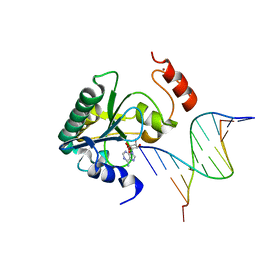 | | Human Aprataxin (Aptx) bound to RNA-DNA and Zn - adenosine vanadate transition state mimic complex | | Descriptor: | 5'-D(*GP*AP*AP*TP*CP*AP*TP*AP*AP*C)-3', 5'-R(P*G)-D(P*TP*TP*AP*TP*GP*AP*TP*TP*C)-3', Aprataxin, ... | | Authors: | Schellenberg, M.J, Tumbale, P.S, Williams, R.S. | | Deposit date: | 2013-10-26 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.541 Å) | | Cite: | Aprataxin resolves adenylated RNA-DNA junctions to maintain genome integrity.
Nature, 506, 2013
|
|
5Z3A
 
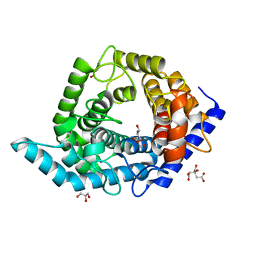 | | Glycosidase Wild Type | | Descriptor: | CITRIC ACID, GLYCEROL, Glycoside hydrolase 15-related protein | | Authors: | Tanaka, Y, Chen, M, Tagami, T, Yao, M, Kimura, A. | | Deposit date: | 2018-01-05 | | Release date: | 2019-05-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Structural insights reveal the second base catalyst of isomaltose glucohydrolase.
Febs J., 289, 2022
|
|
2JPF
 
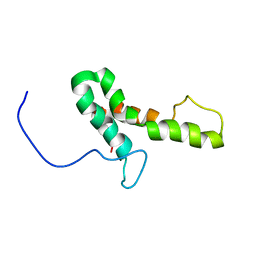 | |
3L4O
 
 | |
5Z9O
 
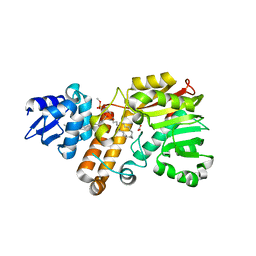 | |
2X0I
 
 | | 2.9 A RESOLUTION STRUCTURE OF MALATE DEHYDROGENASE FROM ARCHAEOGLOBUS FULGIDUS IN COMPLEX WITH NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, MALATE DEHYDROGENASE, SODIUM ION, ... | | Authors: | Irimia, A, Madern, D, Zaccai, G, Vellieux, F.M.D, Karshikoff, A, Tibbelin, G, Ladenstein, R, Lien, T, Birkeland, N.-K. | | Deposit date: | 2009-12-14 | | Release date: | 2009-12-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | The 2.9A Resolution Crystal Structure of Malate Dehydrogenase from Archaeoglobus Fulgidus: Mechanisms of Oligomerisation and Thermal Stabilisation.
J.Mol.Biol., 335, 2004
|
|
3KEA
 
 | |
4S1V
 
 | |
5ZBH
 
 | | The Crystal Structure of Human Neuropeptide Y Y1 Receptor with BMS-193885 | | Descriptor: | Neuropeptide Y receptor type 1,T4 Lysozyme,Neuropeptide Y receptor type 1, dimethyl 4-{3-[({3-[4-(3-methoxyphenyl)piperidin-1-yl]propyl}carbamoyl)amino]phenyl}-2,6-dimethyl-1,4-dihydropyridine-3,5-dicarboxylate | | Authors: | Yang, Z, Han, S, Zhao, Q, Wu, B. | | Deposit date: | 2018-02-11 | | Release date: | 2018-04-25 | | Last modified: | 2018-05-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of ligand binding modes at the neuropeptide Y Y1receptor
Nature, 556, 2018
|
|
6JUJ
 
 | | Crystal structure of Formate dehydrogenase mutant V198I/C256I/P260S/E261P/S381N/S383F from Pseudomonas sp. 101in complex with non-natural cofactor Nicotinamide Cytosine Dinucleotide | | Descriptor: | Formate dehydrogenase, GLYCEROL, [[(2S,3S,4R,5S)-5-(3-aminocarbonylpyridin-1-ium-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2S,3S,4R,5S)-5-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Feng, Y, Guo, X, Xue, S, Zhao, Z. | | Deposit date: | 2019-04-14 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.183 Å) | | Cite: | Structure-Guided Design of Formate Dehydrogenase for Regeneration of a Non-Natural Redox Cofactor.
Chemistry, 26, 2020
|
|
4RX1
 
 | | Crystal Structure of antibiotic-resistance methyltransferase Kmr | | Descriptor: | GLYCEROL, IODIDE ION, Putative rRNA methyltransferase | | Authors: | Savic, M. | | Deposit date: | 2014-12-08 | | Release date: | 2015-03-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.472 Å) | | Cite: | 30S Subunit-Dependent Activation of the Sorangium cellulosum So ce56 Aminoglycoside Resistance-Conferring 16S rRNA Methyltransferase Kmr.
Antimicrob.Agents Chemother., 59, 2015
|
|
