3HCH
 
 | | Structure of the C-terminal domain (MsrB) of Neisseria meningitidis PilB (complex with substrate) | | Descriptor: | (2S)-2-(acetylamino)-N-methyl-4-[(R)-methylsulfinyl]butanamide, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CITRIC ACID, ... | | Authors: | Ranaivoson, F.M, Kauffmann, B, Favier, F. | | Deposit date: | 2009-05-06 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Methionine sulfoxide reductase B displays a high level of flexibility.
J.Mol.Biol., 394, 2009
|
|
3H8W
 
 | |
4NSI
 
 | | Carboplatin binding to HEWL in 20% propanol, 20% PEG 4000 at pH5.6 | | Descriptor: | CITRATE ANION, DIMETHYL SULFOXIDE, Lysozyme C, ... | | Authors: | Tanley, S.W.M, Diederichs, K, Kroon-Batenburg, L.M.J, Levy, C, Schreurs, A.M.M, Helliwell, J.R. | | Deposit date: | 2013-11-28 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Carboplatin binding to histidine.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
6BUV
 
 | | Structure of Mycobacterium tuberculosis NadD in complex with inhibitor [(1~{R},2~{R},5~{S})-5-methyl-2-propan-2-yl-cyclohexyl] 2-[3-methyl-2-(phenoxymethyl)benzimidazol-1-yl]ethanoate | | Descriptor: | 1-methyl-3-(2-{[(1R,2R,5S)-5-methyl-2-(propan-2-yl)cyclohexyl]oxy}-2-oxoethyl)-2-(phenoxymethyl)-1H-1,3-benzimidazol-3-ium, CHLORIDE ION, SODIUM ION, ... | | Authors: | Rodionova, I.A, Reed, R.W, Sorci, L, Osterman, A.L, Korotkov, K.V. | | Deposit date: | 2017-12-11 | | Release date: | 2018-12-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Novel Antimycobacterial Compounds Suppress NAD Biogenesis by Targeting a Unique Pocket of NaMN Adenylyltransferase.
Acs Chem.Biol., 14, 2019
|
|
6H00
 
 | | Crystal structure of human pyridoxine 5-phophate oxidase, R116Q variant | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN MONONUCLEOTIDE, Pyridoxine-5'-phosphate oxidase, ... | | Authors: | Mackinnon, S, Wilson, M.P, Shrestha, L, Bezerra, G.A, Newman, J, Fox, N, Sorrell, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Clayton, P.T, Mills, P.B, Yue, W.W. | | Deposit date: | 2018-07-05 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal structure of human pyridoxine 5-phophate oxidase, R116Q variant
To Be Published
|
|
3EJU
 
 | | Golgi alpha-Mannosidase II in complex with 5-substituted swainsonine analog:(5S)-5-[2'-oxo-2'-(4-tert-butylphenyl)ethyl]-swainsonine | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 1-(4-tert-butylphenyl)-2-[(1S,2R,5S,8R,8aR)-1,2,8-trihydroxyoctahydroindolizin-5-yl]ethanone, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kuntz, D.A, Shea, K, Rose, D.R. | | Deposit date: | 2008-09-18 | | Release date: | 2009-10-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structural Investigation of the Binding of 5-Substituted Swainsonine Analogues to Golgi alpha-Mannosidase II.
Chembiochem, 11, 2010
|
|
2WXU
 
 | | Clostridium perfringens alpha-toxin strain NCTC8237 mutant T74I | | Descriptor: | CADMIUM ION, CALCIUM ION, PHOSPHOLIPASE C, ... | | Authors: | Vachieri, S.G, Naylor, C.E, Basak, A.K. | | Deposit date: | 2009-11-10 | | Release date: | 2009-11-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparison of a Nontoxic Variant of Clostridium Perfringens [Alpha]-Toxin with the Toxic Wild-Type Strain
Acta Crystallogr.,Sect.D, 66, 2010
|
|
6I4Q
 
 | | Crystal structure of the human dihydrolipoamide dehydrogenase at 1.75 Angstrom resolution | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Dihydrolipoyl dehydrogenase, mitochondrial, ... | | Authors: | Nagy, B, Szabo, E, Wilk, P, Torocsik, B, Weiss, M.S, Adam-Vizi, V, Ambrus, A. | | Deposit date: | 2018-11-10 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Underlying molecular alterations in human dihydrolipoamide dehydrogenase deficiency revealed by structural analyses of disease-causing enzyme variants.
Hum.Mol.Genet., 28, 2019
|
|
2WPC
 
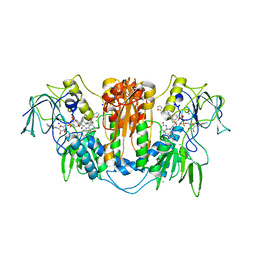 | | Trypanosoma brucei trypanothione reductase in complex with 3,4- dihydroquinazoline inhibitor (DDD00073357) | | Descriptor: | (4S)-6-CHLORO-3-{2-[4-(FURAN-2-YLCARBONYL)PIPERAZIN-1-YL]ETHYL}-2-METHYL-4-PHENYL-3,4-DIHYDROQUINAZOLINE, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Alphey, M.S, Patterson, S, Fairlamb, A.H. | | Deposit date: | 2009-08-05 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Dihydroquinazolines as a Novel Class of Trypanosoma Brucei Trypanothione Reductase Inhibitors: Discovery, Synthesis, and Characterization of Their Binding Mode by Protein Crystallography.
J.Med.Chem., 54, 2011
|
|
4PPO
 
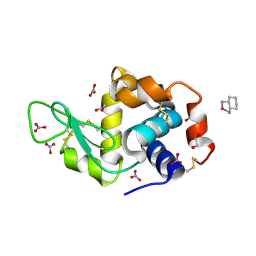 | | First Crystal Structure for an Oxaliplatin-Protein Complex | | Descriptor: | 1,2-ETHANEDIOL, CYCLOHEXANE-1(R),2(R)-DIAMINE-PLATINUM(II), Lysozyme C, ... | | Authors: | Messori, L, Merlino, A. | | Deposit date: | 2014-02-27 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | The X-ray structure of the complex formed in the reaction between oxaliplatin and lysozyme.
Chem.Commun.(Camb.), 50, 2014
|
|
4PPU
 
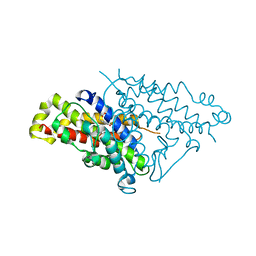 | |
6GII
 
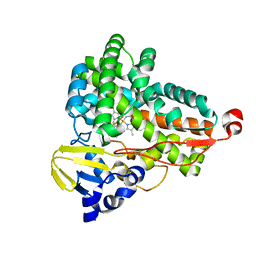 | | The crystal structure of Tepidiphilus thermophilus P450 heme domain | | Descriptor: | Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Levy, C.W. | | Deposit date: | 2018-05-11 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of P450-TT heme-domain provides the first structural insights into the versatile class VII P450s.
Biochem. Biophys. Res. Commun., 501, 2018
|
|
3ETE
 
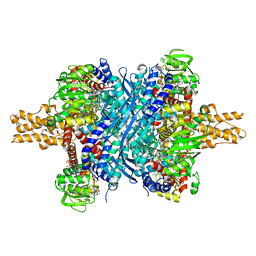 | | Crystal structure of bovine glutamate dehydrogenase complexed with hexachlorophene | | Descriptor: | 2,2'-methanediylbis(3,4,6-trichlorophenol), GLUTAMIC ACID, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Li, M, Smith, T.J. | | Deposit date: | 2008-10-07 | | Release date: | 2009-06-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Novel Inhibitors Complexed with Glutamate Dehydrogenase: ALLOSTERIC REGULATION BY CONTROL OF PROTEIN DYNAMICS
J.Biol.Chem., 284, 2009
|
|
6BUQ
 
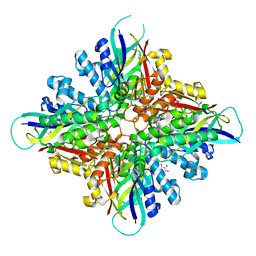 | |
6C4K
 
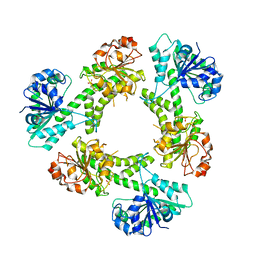 | |
4NWU
 
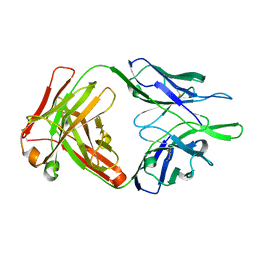 | |
6KA1
 
 | | E.coli Malate dehydrogenase | | Descriptor: | Malate dehydrogenase | | Authors: | Wang, H, Wang, M, Sun, H. | | Deposit date: | 2019-06-19 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.543 Å) | | Cite: | MDH is a major silver target in E. coli
To Be Published
|
|
2X5L
 
 | | X-RAY STRUCTURE OF THE SUBSTRATE-FREE MYCOBACTERIUM TUBERCULOSIS CYTOCHROME P450 CYP125, ALTERNATIVE CRYSTAL FORM | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, PUTATIVE CYTOCHROME P450 125 | | Authors: | Ouellet, H, Kells, P.M, Ortiz de Montellano, P.R, Podust, L.M. | | Deposit date: | 2010-02-10 | | Release date: | 2010-11-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Reverse Type I Inhibitor of Mycobacteriumtuberculosis Cyp125A1.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3GMS
 
 | |
4PE8
 
 | | Crystal structure of TatD in complex with trinucleotide DNA | | Descriptor: | DNA (5'-D(*GP*CP*T)-3'), Tat-linked quality control protein TatD | | Authors: | Chen, Y, Li, C.-L, Hsiao, Y.-Y, Duh, Y, Yuan, H.S. | | Deposit date: | 2014-04-23 | | Release date: | 2014-08-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.894 Å) | | Cite: | Structure and function of TatD exonuclease in DNA repair.
Nucleic Acids Res., 42, 2014
|
|
4NZ3
 
 | | Structure of Vibrio cholerae chitin de-N-acetylase in complex with DI(N-ACETYL-D-GLUCOSAMINE) (CBS) in P 21 21 21 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Deacetylase DA1, ... | | Authors: | Albesa-Jove, D, Andres, E, Biarnes, X, Planas, A, Guerin, M.E. | | Deposit date: | 2013-12-11 | | Release date: | 2014-08-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.114 Å) | | Cite: | Structural basis of chitin oligosaccharide deacetylation.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
6K35
 
 | | Crystal structure of GH20 exo beta-N-acetylglucosaminidase from Vibrio harveyi in complex with NAG-thiazoline | | Descriptor: | 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, Beta-N-acetylglucosaminidase Nag2 | | Authors: | Meekrathok, P, Stubbs, K.A, Bulmer, D.M, van den Berg, B, Suginta, W. | | Deposit date: | 2019-05-16 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | NAG-thiazoline is a potent inhibitor of the Vibrio campbellii GH20 beta-N-Acetylglucosaminidase.
Febs J., 287, 2020
|
|
6GLJ
 
 | | Crystal structure of hMTH1 F27A in complex with TH scaffold 1 in the absence of acetate | | Descriptor: | 4-phenylpyrimidin-2-amine, 7,8-dihydro-8-oxoguanine triphosphatase, SULFATE ION | | Authors: | Eberle, S.A, Wiedmer, L, Sledz, P, Caflisch, A. | | Deposit date: | 2018-05-23 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.301 Å) | | Cite: | hMTH1 F27A in complex with TH scaffold 1.
To Be Published
|
|
3GN0
 
 | |
6GLO
 
 | |
