1RAS
 
 | | CRYSTAL STRUCTURE OF A FLUORESCENT DERIVATIVE OF RNASE A | | Descriptor: | 5-(1-SULFONAPHTHYL)-ACETYLAMINO-ETHYLAMINE, RIBONUCLEASE A | | Authors: | Baudet-Nessler, S, Jullien, M, Crosio, M.-P, Janin, J. | | Deposit date: | 1993-03-02 | | Release date: | 1993-10-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a fluorescent derivative of RNase A.
Biochemistry, 32, 1993
|
|
2H5L
 
 | | S-Adenosylhomocysteine hydrolase containing NAD and 3-deaza-D-eritadenine | | Descriptor: | (2R,3R)-4-(4-AMINO-1H-IMIDAZO[4,5-C]PYRIDIN-1-YL)-2,3-DIHYDROXYBUTANOIC ACID, Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yamada, T, Komoto, J, Takusagawa, F. | | Deposit date: | 2006-05-26 | | Release date: | 2007-04-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and function of eritadenine and its 3-deaza analogues: Potent inhibitors of S-adenosylhomocysteine hydrolase and hypocholesterolemic agents.
Biochem.Pharm., 73, 2007
|
|
2XHS
 
 | |
1RAR
 
 | | CRYSTAL STRUCTURE OF A FLUORESCENT DERIVATIVE OF RNASE A | | Descriptor: | 5-(1-SULFONAPHTHYL)-ACETYLAMINO-ETHYLAMINE, CHLORIDE ION, RIBONUCLEASE A | | Authors: | Baudet-Nessler, S, Jullien, M, Crosio, M.-P, Janin, J. | | Deposit date: | 1993-03-02 | | Release date: | 1993-10-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a fluorescent derivative of RNase A.
Biochemistry, 32, 1993
|
|
2BVB
 
 | | The C-terminal domain from Micronemal Protein 1 (MIC1) from Toxoplasma Gondii | | Descriptor: | MICRONEMAL PROTEIN 1 | | Authors: | Saouros, S, Edwards-Jones, B, Reiss, M, Sawmynaden, K, Cota, E, Simpson, P, Dowse, T.J, Jakle, U, Ramboarina, S, Shivarattan, T, Matthews, S, Soldati-Favre, D. | | Deposit date: | 2005-06-23 | | Release date: | 2005-10-12 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | A Novel Galectin-Like Domain from Toxoplasma Gondll Micronemal Protein 1 Assists the Folding, Assembly,and Transport of a Cell-Adhesion Complex.
J.Biol.Chem., 280, 2005
|
|
7EV2
 
 | |
3UG4
 
 | | Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima arabinose complex | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha-L-arabinofuranosidase, alpha-L-arabinofuranose | | Authors: | Im, D.-H, Miyazaki, K, Wakagi, T, Fushinobu, S. | | Deposit date: | 2011-11-02 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structures of Glycoside Hydrolase Family 51 alpha-L-Arabinofuranosidase from Thermotoga maritima
Biosci.Biotechnol.Biochem., 76, 2012
|
|
3UGX
 
 | | Crystal Structure of Visual Arrestin | | Descriptor: | 1,2-ETHANEDIOL, IMIDAZOLE, PENTANEDIAL, ... | | Authors: | Batra-Safferling, R, Granzin, J. | | Deposit date: | 2011-11-03 | | Release date: | 2012-02-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.649 Å) | | Cite: | Crystal Structure of p44, a Constitutively Active Splice Variant of Visual Arrestin.
J.Mol.Biol., 416, 2012
|
|
4I1U
 
 | |
4QMU
 
 | | MST3 IN COMPLEX WITH JNJ-7706621, 4-({5-AMINO-1-[(2,6-DIFLUOROPHENYL)CARBONYL]-1H-1,2,4-TRIAZOL-3-YL}AMINO)BENZENESULFONAMIDE | | Descriptor: | 4-({5-amino-1-[(2,6-difluorophenyl)carbonyl]-1H-1,2,4-triazol-3-yl}amino)benzenesulfonamide, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Olesen, S.H, Watts, C, Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2014-06-16 | | Release date: | 2015-07-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.546 Å) | | Cite: | Discovery of Diverse Small-Molecule Inhibitors of Mammalian Sterile20-like Kinase 3 (MST3).
Chemmedchem, 11, 2016
|
|
3BWV
 
 | |
1OIV
 
 | | X-ray structure of the small G protein Rab11a in complex with GDP | | Descriptor: | 1,2-ETHANEDIOL, GUANOSINE-5'-DIPHOSPHATE, RAS-RELATED PROTEIN RAB-11A, ... | | Authors: | Pasqualato, S, Senic-Matuglia, F, Renault, L, Goud, B, Salamero, J, Cherfils, J. | | Deposit date: | 2003-06-26 | | Release date: | 2004-01-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The Structural Gdp/GTP Cycle of Rab11 Reveals a Novel Interface Involved in the Dynamics of Recycling Endosomes
J.Biol.Chem., 279, 2004
|
|
5A6M
 
 | | Determining the specificities of the catalytic site from the very high resolution structure of the thermostable glucuronoxylan endo-Beta-1, 4-xylanase, CtXyn30A, from Clostridium thermocellum with a xylotetraose bound | | Descriptor: | CARBOHYDRATE BINDING FAMILY 6, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | Authors: | Freire, F, Verma, A.K, Bule, P, Goyal, A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2015-06-30 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Conservation in the Mechanism of Glucuronoxylan Hydrolysis Revealed by the Structure of Glucuronoxylan Xylano-Hydrolase (Ctxyn30A) from Clostridium Thermocellum
Acta Crystallogr.,Sect.D, 72, 2016
|
|
7K1O
 
 | | Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with Uridine-3',5'-Diphosphate | | Descriptor: | 1,2-ETHANEDIOL, 1-(3,5-di-O-phosphono-alpha-L-xylofuranosyl)pyrimidine-2,4(1H,3H)-dione, Uridylate-specific endoribonuclease | | Authors: | Kim, Y, Maltseva, N, Jedrzejczak, R, Endres, M, Welk, L, Chang, C, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-09-08 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with Uridine-3',5'-Diphosphate
To Be Published
|
|
5OP5
 
 | | Structure of CHK1 10-pt. mutant complex with pyrrolopyrimidine LRRK2 inhibitor | | Descriptor: | 3-[4-(morpholin-4-yl)-7H-pyrrolo[2,3-d]pyrimidin-5-yl]benzonitrile, CHLORIDE ION, Serine/threonine-protein kinase Chk1 | | Authors: | Dokurno, P, Williamson, D.S, Acheson-Dossang, P, Chen, I, Murray, J.B, Shaw, T, Surgenor, A.E. | | Deposit date: | 2017-08-09 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design of Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Crystallographic Surrogate Derived from Checkpoint Kinase 1 (CHK1).
J. Med. Chem., 60, 2017
|
|
7JV7
 
 | | Crystal Structure of the yeast RNA Pol II CTD kinase CTDK-1 complex | | Descriptor: | CITRATE ANION, CTD kinase subunit alpha, CTD kinase subunit beta, ... | | Authors: | Xie, Y, Ren, Y. | | Deposit date: | 2020-08-20 | | Release date: | 2021-01-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.850553 Å) | | Cite: | Structure and activation mechanism of the yeast RNA Pol II CTD kinase CTDK-1 complex.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
3UF4
 
 | |
3UG5
 
 | | Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima xylose complex | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha-L-arabinofuranosidase, beta-D-xylopyranose | | Authors: | Im, D.-H, Miyazaki, K, Wakagi, T, Fushinobu, S. | | Deposit date: | 2011-11-02 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Glycoside Hydrolase Family 51 alpha-L-Arabinofuranosidase from Thermotoga maritima
Biosci.Biotechnol.Biochem., 76, 2012
|
|
2AEH
 
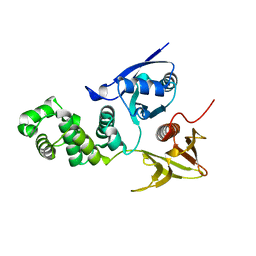 | | Focal adhesion kinase 1 | | Descriptor: | Focal adhesion kinase 1 | | Authors: | Ceccarelli, D.F, Song, H.K, Poy, F, Schaller, M.D, Eck, M.J. | | Deposit date: | 2005-07-22 | | Release date: | 2005-10-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Crystal Structure of the FERM Domain of Focal Adhesion Kinase
J.Biol.Chem., 281, 2006
|
|
2R9R
 
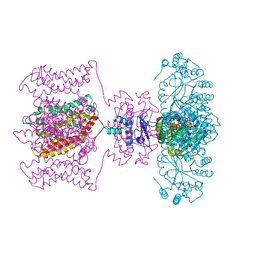 | | Shaker family voltage dependent potassium channel (kv1.2-kv2.1 paddle chimera channel) in association with beta subunit | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, POTASSIUM ION, ... | | Authors: | Long, S.B, Tao, X, Campbell, E.B, Mackinnon, R. | | Deposit date: | 2007-09-13 | | Release date: | 2007-11-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment.
Nature, 450, 2007
|
|
4HSG
 
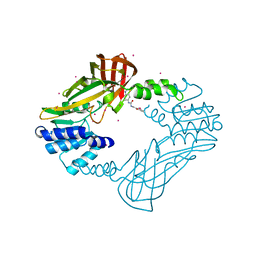 | | Crystal structure of human PRMT3 in complex with an allosteric inhibitor (PRMT3- KTD) | | Descriptor: | 1-(1,2,3-benzothiadiazol-6-yl)-3-(2-oxo-2-phenylethyl)urea, PRMT3 protein, UNKNOWN ATOM OR ION | | Authors: | Dobrovetsky, E, Dong, A, Liu, F, Li, F, Tempel, W, Siarheyeva, A, Hajian, T, Smil, D, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Schapira, M, Jin, J, Vedadi, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-10-30 | | Release date: | 2012-12-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Exploiting an allosteric binding site of PRMT3 yields potent and selective inhibitors.
J. Med. Chem., 56, 2013
|
|
1CL1
 
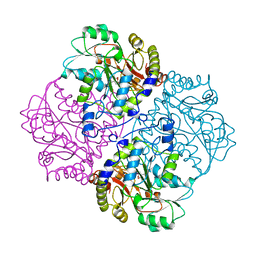 | |
5A94
 
 | | Crystal structure of beta-Glucanase SdGluc5_26A from Saccharophagus degradans in complex with tetrasaccharide A, form 1 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, PUTATIVE RETAINING B-GLYCOSIDASE, ... | | Authors: | Sulzenbacher, G, Lafond, M, Freyd, T, Henrissat, B, Coutinho, R.M, Berrin, J.G, Garron, M.L. | | Deposit date: | 2015-07-17 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The Quaternary Structure of a Glycoside Hydrolase Dictates Specificity Towards Beta-Glucans
J.Biol.Chem., 291, 2016
|
|
5L0E
 
 | | Crystal Structure of Autotaxin and Compound 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-(3-{2-[(2,3-dihydro-1H-inden-2-yl)amino]-7,8-dihydropyrido[4,3-d]pyrimidin-6(5H)-yl}propanoyl)-1,3-benzoxazol-2(3H)-one, CHLORIDE ION, ... | | Authors: | Durbin, J.D. | | Deposit date: | 2016-07-27 | | Release date: | 2016-08-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Novel Autotaxin Inhibitors for the Treatment of Osteoarthritis Pain: Lead Optimization via Structure-Based Drug Design.
Acs Med.Chem.Lett., 7, 2016
|
|
4F76
 
 | |
