9L20
 
 | | cryo-EM structure of Vitamin K-dependent gamma-carboxylase complexed with anisindione | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-(4-methoxyphenyl)-1H-indene-1,3(2H)-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yao, D, Wu, K, Lan, P. | | Deposit date: | 2024-12-16 | | Release date: | 2025-10-22 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | cryo-EM structure of Vitamin K-dependent gamma-carboxylase complexed with anisindione
To Be Published
|
|
9L21
 
 | |
9L24
 
 | |
6LII
 
 | | A quinone oxidoreductase | | Descriptor: | Synaptic vesicle membrane protein VAT-1 homolog | | Authors: | Hakoshima, T, Kim, S.-Y, Mori, T. | | Deposit date: | 2019-12-11 | | Release date: | 2021-01-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into vesicle amine transport-1 (VAT-1) as a member of the NADPH-dependent quinone oxidoreductase family.
Sci Rep, 11, 2021
|
|
1W82
 
 | | p38 Kinase crystal structure in complex with small molecule inhibitor | | Descriptor: | MITOGEN-ACTIVATED PROTEIN KINASE 14, N-[(3Z)-5-TERT-BUTYL-2-PHENYL-1,2-DIHYDRO-3H-PYRAZOL-3-YLIDENE]-N'-(4-CHLOROPHENYL)UREA | | Authors: | Tickle, J, Jhoti, H, Cleasby, A, Devine, L. | | Deposit date: | 2004-09-16 | | Release date: | 2005-02-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of Novel P38Alpha Map Kinase Inhibitors Using Fragment-Based Lead Generation
J.Med.Chem., 48, 2005
|
|
2AFX
 
 | | Crystal structure of human glutaminyl cyclase in complex with 1-benzylimidazole | | Descriptor: | 1-BENZYL-1H-IMIDAZOLE, Glutaminyl-peptide cyclotransferase, SULFATE ION, ... | | Authors: | Huang, K.F, Liu, Y.L, Cheng, W.J, Ko, T.P, Wang, A.H.J. | | Deposit date: | 2005-07-26 | | Release date: | 2005-08-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structures of human glutaminyl cyclase, an enzyme responsible for protein N-terminal pyroglutamate formation
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
4C9S
 
 | | BACTERIAL CHALCONE ISOMERASE IN open CONFORMATION FROM EUBACTERIUM RAMULUS AT 1.8 A RESOLUTION | | Descriptor: | CHALCONE ISOMERASE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Thomsen, M, Palm, G.J, Hinrichs, W. | | Deposit date: | 2013-10-03 | | Release date: | 2014-10-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and Catalytic Mechanism of the Evolutionarily Unique Bacterial Chalcone Isomerase
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1MVY
 
 | | Amylosucrase mutant E328Q co-crystallized with maltoheptaose. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Skov, L.K, Mirza, O, Sprogoe, D, Dar, I, Remaud-Simeon, M, Albenne, C, Monsan, P, Gajhede, M. | | Deposit date: | 2002-09-27 | | Release date: | 2002-12-18 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Oligosaccharide and Sucrose Complexes of Amylosucrase. STRUCTURAL IMPLICATIONS FOR THE POLYMERASE ACTIVITY
J.BIOL.CHEM., 277, 2002
|
|
2O40
 
 | |
4NR5
 
 | | Crystal structure of the bromodomain of human CREBBP in complex with an isoxazolyl-benzimidazole ligand | | Descriptor: | 1,2-ETHANEDIOL, 5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[2-(morpholin-4-yl)ethyl]-2-(2-phenylethyl)-1H-benzimidazole, CREB-binding protein, ... | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Hay, D, Fedorov, O, Martin, S, Pike, A.W, von Delft, F, Brennan, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-11-26 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal structure of the bromodomain of human CREBBP in complex with an isoxazolyl-benzimidazole ligand
To be Published
|
|
4C0M
 
 | | Crystal Structure of the N terminal domain of wild type TRIF (TIR- domain-containing adapter-inducing interferon-beta) | | Descriptor: | TIR DOMAIN-CONTAINING ADAPTER MOLECULE 1 | | Authors: | Ullah, M.O, Ve, T, Mangan, M, Alaidarous, M, Sweet, M.J, Mansell, A, Kobe, B. | | Deposit date: | 2013-08-05 | | Release date: | 2013-12-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Tlr Signalling Adaptor Trif/Ticam-1 Has an N-Terminal Helical Domain with Structural Similarity to Ifit Proteins
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1QXM
 
 | | Crystal structure of a hemagglutinin component (HA1) from type C Clostridium botulinum | | Descriptor: | 1,2-ETHANEDIOL, HA1 | | Authors: | Inoue, K, Sobhany, M, Transue, T.R, Oguma, K, Pedersen, L.C, Negishi, M. | | Deposit date: | 2003-09-08 | | Release date: | 2004-01-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural analysis by X-ray crystallography and calorimetry of a haemagglutinin component (HA1) of the progenitor toxin from Clostridium botulinum.
Microbiology, 149, 2003
|
|
4C1C
 
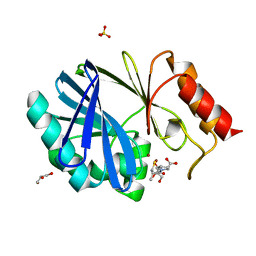 | | Crystal structure of the metallo-beta-lactamase BCII with D-captopril | | Descriptor: | 1-(3-MERCAPTO-2-METHYL-PROPIONYL)-PYRROLIDINE-2-CARBOXYLIC ACID, BETA-LACTAMASE 2, GLYCEROL, ... | | Authors: | Zollman, D, Brem, J, McDonough, M.A, van Berkel, S.S, Schofield, C.J. | | Deposit date: | 2013-08-12 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Structural Basis of Metallo-beta-Lactamase Inhibition by Captopril Stereoisomers.
Antimicrob. Agents Chemother., 60, 2015
|
|
3N25
 
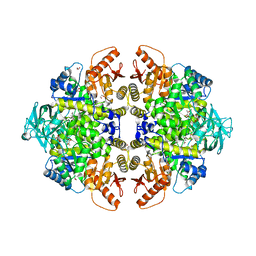 | | The structure of muscle pyruvate kinase in complex with proline, pyruvate, and Mn2+ | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Fenton, A.W, Johnson, T.A, Holyoak, T. | | Deposit date: | 2010-05-17 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | The pyruvate kinase model system, a cautionary tale for the use of osmolyte perturbations to support conformational equilibria in allostery.
Protein Sci., 19, 2010
|
|
3UYS
 
 | | Crystal structure of apo human ck1d | | Descriptor: | Casein kinase I isoform delta, SULFATE ION | | Authors: | Huang, X. | | Deposit date: | 2011-12-06 | | Release date: | 2012-01-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the interaction between casein kinase 1 delta and a potent and selective inhibitor.
J.Med.Chem., 55, 2012
|
|
2O6E
 
 | | Structure of native rTp34 from Treponema pallidum from zinc-soaked crystals | | Descriptor: | 1,2-ETHANEDIOL, 34 kDa membrane antigen, CHLORIDE ION, ... | | Authors: | Machius, M, Brautigam, C.A, Deka, R.K, Tomchick, D.R, Lumpkins, S.B, Norgard, M.V. | | Deposit date: | 2006-12-07 | | Release date: | 2006-12-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the Tp34 (TP0971) lipoprotein of treponema pallidum: implications of its metal-bound state and affinity for human lactoferrin.
J.Biol.Chem., 282, 2007
|
|
7GTF
 
 | | PanDDA Analysis group deposition -- Crystal structure of PTP1B in complex with XST00000754b | | Descriptor: | 1,2-benzoxazol-3-ol, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Ginn, H.M, Keedy, D.A. | | Deposit date: | 2024-01-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | An expanded view of ligandability in the allosteric enzyme PTP1B from computational reanalysis of large-scale crystallographic data.
Biorxiv, 2024
|
|
3DX1
 
 | | Golgi alpha-Mannosidase II in complex with Mannostatin analog (1S,2S,3R,4R)-4-aminocyclopentane-1,2,3-triol | | Descriptor: | (1S,2S,3R,4R)-4-aminocyclopentane-1,2,3-triol, (4R)-2-METHYLPENTANE-2,4-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kuntz, D.A, Rose, D.R. | | Deposit date: | 2008-07-23 | | Release date: | 2009-07-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | The molecular basis of inhibition of Golgi alpha-mannosidase II by mannostatin A.
Chembiochem, 10, 2009
|
|
7GSK
 
 | |
4NRY
 
 | |
4C4A
 
 | | Crystal structure of mouse protein arginine methyltransferase 7 in complex with SAH | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 3-[BENZYL(DIMETHYL)AMMONIO]PROPANE-1-SULFONATE, PROTEIN ARGININE N-METHYLTRANSFERASE 7, ... | | Authors: | Cura, V, Troffer-Charlier, N, Bonnefond, L, Wurtz, J.M, Cavarelli, J. | | Deposit date: | 2013-09-02 | | Release date: | 2014-07-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Insight Into Arginine Methylation by the Mouse Protein Arginine Methyltransferase 7: A Zinc Finger Freezes the Mimic of the Dimeric State Into a Single Active Site.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6KVV
 
 | |
4CAF
 
 | | Plasmodium vivax N-myristoyltransferase in complex with a benzothiophene inhibitor (compound 34a) | | Descriptor: | 2-oxopentadecyl-CoA, 4-[(2-{5-[(3,5-dimethyl-1H-pyrazol-4-yl)methyl]-1,3,4-oxadiazol-2-yl}-1-benzothiophen-3-yl)oxy]piperidine, AMMONIUM ION, ... | | Authors: | Rackham, M.D, Brannigan, J.A, Rangachari, K, Wilkinson, A.J, Holder, A.A, Tate, E.W, Leatherbarrow, R.J. | | Deposit date: | 2013-10-08 | | Release date: | 2014-04-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design and Synthesis of High Affinity Inhibitors of Plasmodium Falciparum and Plasmodium Vivax N-Myristoyltransferases Directed by Ligand Efficiency Dependent Lipophilicity (Lelp).
J.Med.Chem., 57, 2014
|
|
2PTW
 
 | | Crystal Structure of the T. brucei enolase complexed with sulphate, identification of a metal binding site IV | | Descriptor: | 1,2-ETHANEDIOL, Enolase, SULFATE ION, ... | | Authors: | Navarro, M.V.A.S, Rigden, D.J, Garratt, R.C, Dias, S.M.G. | | Deposit date: | 2007-05-08 | | Release date: | 2007-11-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural flexibility in Trypanosoma brucei enolase revealed by X-ray crystallography and molecular dynamics.
Febs J., 274, 2007
|
|
7G1M
 
 | | Crystal Structure of human FABP4 binding site mutated to that of FABP5 in complex with rac-(1R,2R)-2-[[3-(3-methyl-1,2,4-oxadiazol-5-yl)-4,5,6,7-tetrahydro-1-benzothiophen-2-yl]carbamoyl]cyclohexane-1-carboxylic acid, i.e. SMILES C1(=C(C2=C(S1)CCCC2)C1=NC(=NO1)C)NC(=O)[C@@H]1[C@H](CCCC1)C(=O)O with IC50=0.365 microM | | Descriptor: | (1R,2S)-2-{[(3M)-3-(3-methyl-1,2,4-oxadiazol-5-yl)-4,5,6,7-tetrahydro-1-benzothiophen-2-yl]carbamoyl}cyclohexane-1-carboxylic acid, Fatty acid-binding protein, adipocyte | | Authors: | Ehler, A, Benz, J, Obst, U, Neidhart, W, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
