2ZDA
 
 | | Exploring Thrombin S1 pocket | | Descriptor: | D-phenylalanyl-N-{4-[amino(iminio)methyl]benzyl}-L-prolinamide, Hirudin variant-1, PHOSPHATE ION, ... | | Authors: | Baum, B, Heine, A, Klebe, G. | | Deposit date: | 2007-11-21 | | Release date: | 2008-10-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Think twice: understanding the high potency of bis(phenyl)methane inhibitors of thrombin
J.Mol.Biol., 391, 2009
|
|
7SZV
 
 | |
7DXS
 
 | |
3HOM
 
 | |
7PPG
 
 | | CRYSTAL STRUCTURE OF NAMPT IN COMPLEX WITH COMPOUND 9 | | Descriptor: | 1,2-ETHANEDIOL, N-[4-[(4R)-1-cyclopentyl-4-methyl-6-oxidanylidene-4,5-dihydropyridazin-3-yl]phenyl]-1,3-dihydropyrrolo[3,4-c]pyridine-2-carboxamide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Hillig, R.C. | | Deposit date: | 2021-09-13 | | Release date: | 2022-06-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | A Novel NAMPT Inhibitor-Based Antibody-Drug Conjugate Payload Class for Cancer Therapy.
Bioconjug.Chem., 33, 2022
|
|
7PXM
 
 | | X-ray structure of LPMO at 1.45x10^6 Gy | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Auxiliary activity 9, COPPER (II) ION | | Authors: | Tandrup, T, Lo Leggio, L. | | Deposit date: | 2021-10-08 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
2ZDO
 
 | | Crystal structure of IsdG-N7A in complex with hemin | | Descriptor: | Heme-degrading monooxygenase isdG, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lee, W.C, Reniere, M.L, Skaar, E.P, Murphy, M.E.P. | | Deposit date: | 2007-11-27 | | Release date: | 2008-08-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ruffling of Metalloporphyrins Bound to IsdG and IsdI, Two Heme-degrading Enzymes in Staphylococcus aureus
J.Biol.Chem., 283, 2008
|
|
5KR7
 
 | | KDM4C bound to pyrazolo-pyrimidine scaffold | | Descriptor: | 6-ethyl-2,5-dimethyl-7-oxidanylidene-4~{H}-pyrazolo[1,5-a]pyrimidine-3-carbonitrile, CHLORIDE ION, FE (II) ION, ... | | Authors: | Bellon, S.F, Poy, F, Setser, J.W. | | Deposit date: | 2016-07-07 | | Release date: | 2016-08-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of potent, selective KDM5 inhibitors.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
7SZ1
 
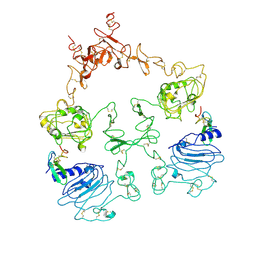 | | Cryo-EM structure of the extracellular module of the full-length EGFR L834R bound to EGF. "tips-separated" conformation | | Descriptor: | Epidermal growth factor, Epidermal growth factor receptor | | Authors: | Huang, Y, Ognjenovic, J, Karandur, D, Miller, K, Merk, A, Subramaniam, S, Kuriyan, J. | | Deposit date: | 2021-11-25 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | A molecular mechanism for the generation of ligand-dependent differential outputs by the epidermal growth factor receptor.
Elife, 10, 2021
|
|
7B6Q
 
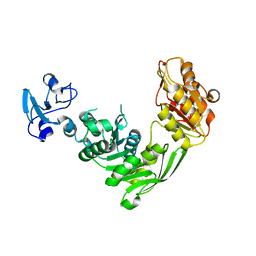 | | Crystal structure of MurE from E.coli in complex with Z57299526 | | Descriptor: | ISOPROPYL ALCOHOL, N-[(furan-2-yl)methyl]-1H-benzimidazol-2-amine, UDP-N-acetylmuramoyl-L-alanyl-D-glutamate-2,6-diaminopimelate ligase | | Authors: | Koekemoer, L, Steindel, M, Fairhead, M, Talon, R, Douangamath, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Krojer, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-08 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of MurE from E.coli
To Be Published
|
|
7SYD
 
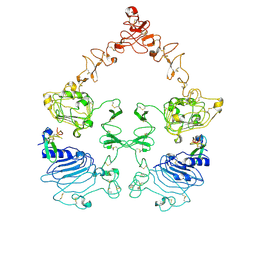 | | Cryo-EM structure of the extracellular module of the full-length EGFR bound to EGF "tips-juxtaposed" conformation | | Descriptor: | Epidermal growth factor, Epidermal growth factor receptor | | Authors: | Huang, Y, Ognjenovic, J, Karandur, D, Miller, K, Merk, A, Subramaniam, S, Kuriyan, J. | | Deposit date: | 2021-11-24 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A molecular mechanism for the generation of ligand-dependent differential outputs by the epidermal growth factor receptor.
Elife, 10, 2021
|
|
3HPT
 
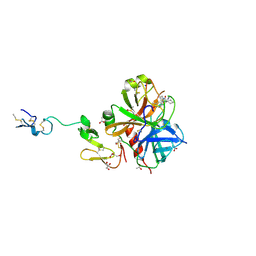 | | Crystal structure of human FxA in complex with (S)-2-cyano-1-(2-methylbenzofuran-5-yl)-3-(2-oxo-1-(2-oxo-2-(pyrrolidin-1-yl)ethyl)azepan-3-yl)guanidine | | Descriptor: | 1-cyano-2-(2-methyl-1-benzofuran-5-yl)-3-[(3S)-2-oxo-1-(2-oxo-2-pyrrolidin-1-ylethyl)azepan-3-yl]guanidine, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Klei, H.E, Ghosh, K, Rushith, A, Kish, K. | | Deposit date: | 2009-06-04 | | Release date: | 2009-11-17 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Cyanoguanidine-based lactam derivatives as a novel class of orally bioavailable factor Xa inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
7JTA
 
 | | Crystal structure of a putative nuclease with anti-Cas9 activity from an uncultured Clostridia bacterium | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, NITRATE ION, NTF2-like nuclease/anti-CRISPR | | Authors: | Werther, R, Forsberg, K.J, Stoddard, B.L. | | Deposit date: | 2020-08-17 | | Release date: | 2021-09-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | The novel anti-CRISPR AcrIIA22 relieves DNA torsion in target plasmids and impairs SpyCas9 activity.
Plos Biol., 19, 2021
|
|
7T6Y
 
 | | d((CGA)5TGA) parallel-stranded homo-duplex | | Descriptor: | BARIUM ION, DNA (5'-D(*CP*GP*AP*CP*GP*AP*CP*GP*AP*CP*GP*AP*CP*GP*AP*TP*GP*A)-3') | | Authors: | Luteran, E.M, Paukstelis, P.J. | | Deposit date: | 2021-12-14 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The parallel-stranded d(CGA) duplex is a highly predictable structural motif with two conformationally distinct strands.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7AO4
 
 | | Crystal structure of CotB2 variant W288G | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Cyclooctat-9-en-7-ol synthase | | Authors: | Dimos, N, Driller, R, Loll, B. | | Deposit date: | 2020-10-13 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Impression of a Nonexisting Catalytic Effect: The Role of CotB2 in Guiding the Complex Biosynthesis of Cyclooctat-9-en-7-ol.
J.Am.Chem.Soc., 142, 2020
|
|
5KSQ
 
 | | Stationary phase survival protein E (SurE) from Xylella fastidiosa | | Descriptor: | 5'-nucleotidase SurE, IODIDE ION, MANGANESE (II) ION, ... | | Authors: | Machado, A.T.P, Fonseca, E.M.B, Dos Reis, M.A, Saraiva, A.M, Dos Santos, C.A, De Toledo, M.A, Polikarpov, I, De Souza, A.P, Aparicio, R, Iulek, J. | | Deposit date: | 2016-07-09 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Conformational variability of the stationary phase survival protein E from Xylella fastidiosa revealed by X-ray crystallography, small-angle X-ray scattering studies, and normal mode analysis.
Proteins, 85, 2017
|
|
2LXY
 
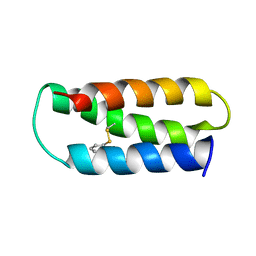 | | NMR structure of 2-MERCAPTOPHENOL-ALPHA3C | | Descriptor: | 2-MERCAPTOPHENOL, 2-mercaptophenol-alpha3C | | Authors: | Tommos, C, Valentine, K.G, Martinez-Rivera, M.C, Liang, L, Moorman, V.R. | | Deposit date: | 2012-09-06 | | Release date: | 2013-02-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Reversible phenol oxidation and reduction in the structurally well-defined 2-Mercaptophenol-alpha(3)C protein.
Biochemistry, 52, 2013
|
|
7DVF
 
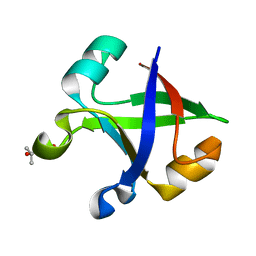 | | Crystal structure of the computationally designed reDPBB_sym2 protein | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, reDPBB_sym2 protein | | Authors: | Yagi, S, Tagami, S, Padhi, A.K, Zhang, K.Y.J. | | Deposit date: | 2021-01-13 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.209 Å) | | Cite: | Seven Amino Acid Types Suffice to Create the Core Fold of RNA Polymerase.
J.Am.Chem.Soc., 143, 2021
|
|
7SYE
 
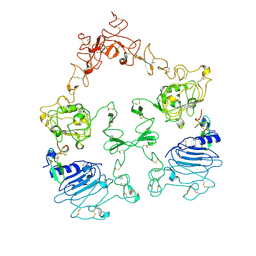 | | Cryo-EM structure of the extracellular module of the full-length EGFR bound to EGF. "tips-separated" conformation | | Descriptor: | Epidermal growth factor, Epidermal growth factor receptor | | Authors: | Huang, Y, Ognjenovic, J, Karandur, D, Miller, K, Merk, A, Subramaniam, S, Kuriyan, J. | | Deposit date: | 2021-11-24 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A molecular mechanism for the generation of ligand-dependent differential outputs by the epidermal growth factor receptor.
Elife, 10, 2021
|
|
3HSR
 
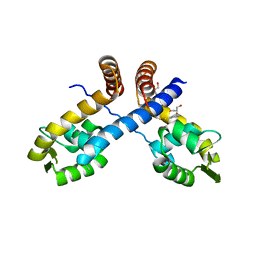 | | Crystal structure of Staphylococcus aureus protein SarZ in mixed disulfide form | | Descriptor: | ACETATE ION, GLYCEROL, HTH-type transcriptional regulator sarZ, ... | | Authors: | Poor, C.B, Duguid, E, Rice, P.A, He, C. | | Deposit date: | 2009-06-10 | | Release date: | 2009-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of the reduced, sulfenic acid, and mixed disulfide forms of SarZ, a redox active global regulator in Staphylococcus aureus.
J.Biol.Chem., 284, 2009
|
|
2ZG7
 
 | | Crystal Structure of Pd(allyl)/apo-Fr | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, Ferritin light chain, ... | | Authors: | Abe, S, Niemeyer, J, Abe, M, Ueno, T, Hikage, T, Erker, G, Watanabe, Y. | | Deposit date: | 2008-01-18 | | Release date: | 2008-08-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Control of the coordination structure of organometallic palladium complexes in an apo-ferritin cage.
J.Am.Chem.Soc., 130, 2008
|
|
3HRP
 
 | |
2ZT7
 
 | |
3HSZ
 
 | | Crystal structure of E. coli HPPK(F123A) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Blaszczyk, J, Li, Y, Yan, H, Ji, X. | | Deposit date: | 2009-06-11 | | Release date: | 2010-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Pterin-binding site mutation Y53A, N55A or F123A and activity of E. coli HPPK
To be Published
|
|
7KE0
 
 | |
