1L9A
 
 | | CRYSTAL STRUCTURE OF SRP19 IN COMPLEX WITH THE S DOMAIN OF SIGNAL RECOGNITION PARTICLE RNA | | Descriptor: | MAGNESIUM ION, METHYL MERCURY ION, SIGNAL RECOGNITION PARTICLE 19 KDA PROTEIN, ... | | Authors: | Oubridge, C, Kuglstatter, A, Jovine, L, Nagai, K. | | Deposit date: | 2002-03-22 | | Release date: | 2002-06-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of SRP19 in complex with the S domain of SRP RNA and its implication for the assembly of the signal recognition particle.
Mol.Cell, 9, 2002
|
|
1L9K
 
 | | dengue methyltransferase | | Descriptor: | RNA-DIRECTED RNA POLYMERASE, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION | | Authors: | Egloff, M.P, Benarroch, D, Selisko, B, Romette, J.L, Canard, B. | | Deposit date: | 2002-03-25 | | Release date: | 2003-03-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An RNA cap (nucleoside-2'-O-) methyltransferase in the flavivirus RNA polymerase NS5: crystal structure and functional characterization
Embo J., 21, 2002
|
|
1LL5
 
 | |
3KER
 
 | |
4E4J
 
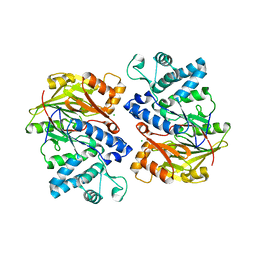 | | Crystal structure of arginine deiminase from Mycoplasma penetrans | | Descriptor: | Arginine deiminase, CHLORIDE ION | | Authors: | Benach, J, Gallego, P, Planell, R, Querol, E, Perez Pons, J.A, Reverter, D. | | Deposit date: | 2012-03-13 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Characterization of the Enzymes Composing the Arginine Deiminase Pathway in Mycoplasma penetrans.
Plos One, 7, 2012
|
|
3QYW
 
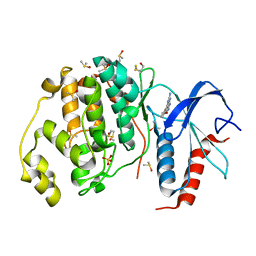 | | Crystal structure of ERK2 in complex with an inhibitor | | Descriptor: | 6-(3-bromophenyl)-7H-purin-2-amine, DIMETHYL SULFOXIDE, Mitogen-activated protein kinase 1, ... | | Authors: | Gelin, M, Pochet, S, Hoh, F, Pirochi, M, Guichou, J.-F, Ferrer, J.-L, Labesse, G. | | Deposit date: | 2011-03-04 | | Release date: | 2011-08-24 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | In-plate protein crystallization, in situ ligand soaking and X-ray diffraction.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
1U9L
 
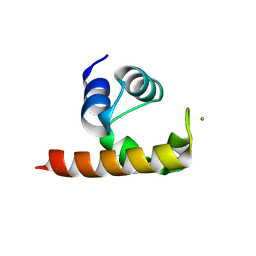 | | Structural basis for a NusA- protein N interaction | | Descriptor: | GOLD ION, Lambda N, Transcription elongation protein nusA | | Authors: | Bonin, I, Muehlberger, R, Bourenkov, G.P, Huber, R, Bacher, A, Richter, G, Wahl, M.C. | | Deposit date: | 2004-08-10 | | Release date: | 2004-08-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the interaction of Escherichia coli NusA with protein N of phage lambda
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1UGM
 
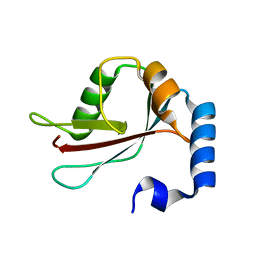 | | Crystal Structure of LC3 | | Descriptor: | Microtubule-associated proteins 1A/1B light chain 3 | | Authors: | Sugawara, K, Suzuki, N.N, Fujioka, Y, Mizushima, N, Ohsumi, Y, Inagaki, F. | | Deposit date: | 2003-06-16 | | Release date: | 2004-07-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The crystal structure of microtubule-associated protein light chain 3, a mammalian homologue of Saccharomyces cerevisiae Atg8
Genes Cells, 9, 2004
|
|
1L8F
 
 | |
3KAV
 
 | | Crystal Structure of THE MUTANT (L80M) PA2107 PROTEIN from Pseudomonas aeruginosa, Northeast Structural Genomics Consortium Target PaR198 | | Descriptor: | uncharacterized protein PA2107 | | Authors: | Forouhar, F, Neely, H, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Foote, E.L, Zhao, L, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-10-19 | | Release date: | 2009-10-27 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Northeast Structural Genomics Consortium Target PaR198
To be Published
|
|
1LAJ
 
 | | The Structure of Tomato Aspermy Virus by X-Ray Crystallography | | Descriptor: | 5'-R(*AP*AP*A)-3', MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Lucas, R.W, Larson, S.B, Canady, M.A, McPherson, A. | | Deposit date: | 2002-03-28 | | Release date: | 2002-11-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The Structure of Tomato Aspermy Virus by X-Ray Crystallography
J.STRUCT.BIOL., 139, 2002
|
|
1TVV
 
 | |
6UMP
 
 | | Crystal structure of MavC in complex with substrate mimic in P65 space group | | Descriptor: | MavC, Ubiquitin, Ubiquitin-conjugating enzyme E2 N | | Authors: | Puvar, K, Iyer, S, Luo, Z.Q, Das, C. | | Deposit date: | 2019-10-10 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Legionella effector MavC targets the Ube2N~Ub conjugate for noncanonical ubiquitination.
Nat Commun, 11, 2020
|
|
1UAX
 
 | |
1TVW
 
 | |
1LS6
 
 | | Human SULT1A1 complexed with PAP and p-Nitrophenol | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, P-NITROPHENOL, aryl sulfotransferase | | Authors: | Gamage, N.U, Barnett, A.C, Tresillian, M, Latham, C.F, Liyou, N.E, McManus, M.E, Martin, J.L. | | Deposit date: | 2002-05-17 | | Release date: | 2003-08-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a human carcinogen-converting enzyme, SULT1A1. Structural and kinetic implications of substrate inhibition.
J.Biol.Chem., 278, 2003
|
|
3SY2
 
 | |
5FM7
 
 | | Double-heterohexameric rings of full-length Rvb1(ADP)Rvb2(ADP) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, RVB1, RVB2 | | Authors: | Silva-Martin, N, Dauden, M.I, Glatt, S, Hoffmann, N.A, Mueller, C.W. | | Deposit date: | 2015-11-02 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | The Combination of X-Ray Crystallography and Cryo-Electron Microscopy Provides Insight Into the Overall Architecture of the Dodecameric Rvb1/Rvb2 Complex.
Plos One, 11, 2016
|
|
1UHA
 
 | | Crystal Structure of Pokeweed Lectin-D2 | | Descriptor: | CALCIUM ION, lectin-D2 | | Authors: | Fujii, T, Hayashida, M, Hamasu, M, Ishiguro, M, Hata, Y. | | Deposit date: | 2003-06-27 | | Release date: | 2004-04-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of two lectins from the roots of pokeweed (Phytolacca americana).
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1KQS
 
 | | The Haloarcula marismortui 50S Complexed with a Pretranslocational Intermediate in Protein Synthesis | | Descriptor: | 23S RRNA, 5S RRNA, 6-AMINOHEXANOIC ACID, ... | | Authors: | Schmeing, T.M, Seila, A.C, Hansen, J.L, Freeborn, B, Soukup, J.K, Scaringe, S.A, Strobel, S.A, Moore, P.B, Steitz, T.A. | | Deposit date: | 2002-01-07 | | Release date: | 2002-02-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A pre-translocational intermediate in protein synthesis observed in crystals of enzymatically active 50S subunits.
Nat.Struct.Biol., 9, 2002
|
|
1ULN
 
 | | Crystal Structure of Pokeweed Lectin-D1 | | Descriptor: | lectin-D | | Authors: | Fujii, T, Hayashida, M, Hamasu, M, Ishiguro, M, Hata, Y. | | Deposit date: | 2003-09-16 | | Release date: | 2004-04-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structures of two lectins from the roots of pokeweed (Phytolacca americana).
Acta Crystallogr.,Sect.D, 60, 2004
|
|
3JQ4
 
 | | The structure of the complex of the large ribosomal subunit from D. Radiodurans with the antibiotic lankacidin | | Descriptor: | 23S ribosomal RNA, 5S ribosomal RNA, N-[(1S,2R,3E,5E,7S,9E,11E,13S,15R,19R)-7,13-dihydroxy-1,4,10,19-tetramethyl-17,18-dioxo-16-oxabicyclo[13.2.2]nonadeca-3,5,9,11-tetraen-2-yl]-2-oxopropanamide | | Authors: | Auerbach-Nevo, T, Mermershtain, I, Davidovich, C, Bashan, A, Rozenberg, H, Yonath, A. | | Deposit date: | 2009-09-06 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.52 Å) | | Cite: | The structure of ribosome-lankacidin complex reveals ribosomal sites for synergistic antibiotics
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1UJ2
 
 | | Crystal structure of human uridine-cytidine kinase 2 complexed with products, CMP and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CYTIDINE-5'-MONOPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Suzuki, N.N, Koizumi, K, Fukushima, M, Matsuda, A, Inagaki, F. | | Deposit date: | 2003-07-25 | | Release date: | 2004-05-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the specificity, catalysis, and regulation of human uridine-cytidine kinase
STRUCTURE, 12, 2004
|
|
4DEA
 
 | | Aurora A in complex with YL1-038-18 | | Descriptor: | 1,2-ETHANEDIOL, 4,4'-(pyrimidine-2,4-diyldiimino)dibenzoic acid, Aurora kinase A | | Authors: | Martin, M.P, Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2012-01-20 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Development of o-Chlorophenyl Substituted Pyrimidines as Exceptionally Potent Aurora Kinase Inhibitors.
J.Med.Chem., 55, 2012
|
|
3K17
 
 | |
