4O3M
 
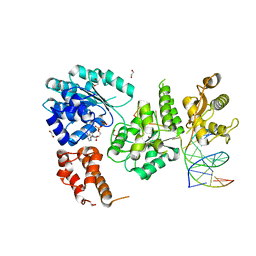 | | Ternary complex of Bloom's syndrome helicase | | Descriptor: | 1,2-ETHANEDIOL, 5'-D(*AP*GP*CP*GP*TP*CP*GP*AP*GP*AP*TP*CP*CP*AP*AP*G)-3', 5'-D(*CP*TP*TP*GP*GP*AP*TP*CP*TP*CP*GP*AP*CP*GP*CP*TP*CP*TP*CP*CP*CP*TP*TP*A)-3', ... | | Authors: | Swan, M.K, Bertrand, J. | | Deposit date: | 2013-12-18 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of human Bloom's syndrome helicase in complex with ADP and duplex DNA.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4BW4
 
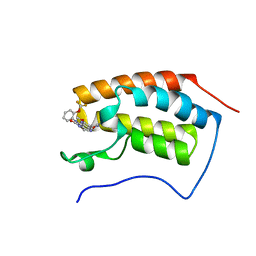 | | The first bromodomain of human BRD4 in complex with 3,5 dimethylisoxaxole ligand | | Descriptor: | 7-(3,5-dimethylisoxazol-4-yl)-8-methoxy-1-(2-(trifluoromethoxy)phenyl)-1h-imidazo[4,5-c][1,5]naphthyridin-2(3h)-one, BROMODOMAIN-CONTAINING PROTEIN 4 | | Authors: | Chung, C, Mirguet, O, Lamotte, Y, Bamborough, P, Delannee, D, Bouillot, A, Gellibert, F, Krysa, G, Lewis, A, Witherington, J, Huet, P, Dudit, Y, Trottet, L, Nicodeme, E. | | Deposit date: | 2013-06-29 | | Release date: | 2013-09-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Naphthyridines as Novel Bet Family Bromodomain Inhibitors.
Chemmedchem, 9, 2014
|
|
4O63
 
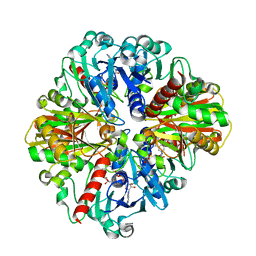 | | Co-enzyme Induced Conformational Changes in Bovine Eye Glyceraldehyde 3-Phosphate Dehydrogenase | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Baker, B.Y, Shi, W, Wang, B, Palczewski, K. | | Deposit date: | 2013-12-20 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | High-resolution crystal structures of the photoreceptor glyceraldehyde 3-phosphate dehydrogenase (GAPDH) with three and four-bound NAD molecules.
Protein Sci., 23, 2014
|
|
4BW2
 
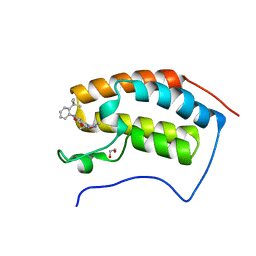 | | The first bromodomain of human BRD4 in complex with 3,5 dimethylisoxaxole ligand | | Descriptor: | 1,2-ETHANEDIOL, 4-((2-(TERT-BUTYL)PHENYL)AMINO)-7-(3,5-dimethylisoxazol-4-yl)-1,8-naphthyridine-3-carboxylic acid, BROMODOMAIN-CONTAINING PROTEIN 4 | | Authors: | Chung, C, Mirguet, O, Lamotte, Y, Bamborough, P, Delannee, D, Bouillot, A, Gellibert, F, Krysa, G, Lewis, A, Witherington, J, Huet, P, Dudit, Y, Trottet, L, Nicodeme, E. | | Deposit date: | 2013-06-29 | | Release date: | 2013-09-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Naphthyridines as Novel Bet Family Bromodomain Inhibitors.
Chemmedchem, 9, 2014
|
|
5NOR
 
 | | Structure of cyclophilin A in complex with 3-methylpyridin-2-amine | | Descriptor: | 3-methylpyridin-2-amine, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Georgiou, C, Mcnae, I.W, Ioannidis, H, Julien, M, Walkinshaw, M.D. | | Deposit date: | 2017-04-13 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Pushing the Limits of Detection of Weak Binding Using Fragment-Based Drug Discovery: Identification of New Cyclophilin Binders.
J. Mol. Biol., 429, 2017
|
|
5NOY
 
 | | Structure of cyclophilin A in complex with 3,4-diaminobenzamide | | Descriptor: | 3,4-bis(azanyl)benzamide, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Georgiou, C, Mcnae, I.W, Ioannidis, H, Julien, M, Walkinshaw, M.D. | | Deposit date: | 2017-04-13 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Pushing the Limits of Detection of Weak Binding Using Fragment-Based Drug Discovery: Identification of New Cyclophilin Binders.
J. Mol. Biol., 429, 2017
|
|
4O70
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with DINACICLIB | | Descriptor: | 1,2-ETHANEDIOL, 3-[({3-ethyl-5-[(2S)-2-(2-hydroxyethyl)piperidin-1-yl]pyrazolo[1,5-a]pyrimidin-7-yl}amino)methyl]-1-hydroxypyridinium, Bromodomain-containing protein 4 | | Authors: | Ember, S.W, Zhu, J.-Y, Watts, C, Schonbrunn, E. | | Deposit date: | 2013-12-24 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Acetyl-lysine Binding Site of Bromodomain-Containing Protein 4 (BRD4) Interacts with Diverse Kinase Inhibitors.
Acs Chem.Biol., 9, 2014
|
|
4O7F
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with SB-251527 | | Descriptor: | 1,2-ETHANEDIOL, 4-[4-(4-fluorophenyl)-1-(piperidin-4-yl)-1H-imidazol-5-yl]-2-(2-methoxyphenoxy)pyrimidine, Bromodomain-containing protein 4, ... | | Authors: | Ember, S.W, Zhu, J.-Y, Watts, C, Schonbrunn, E. | | Deposit date: | 2013-12-24 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Acetyl-lysine Binding Site of Bromodomain-Containing Protein 4 (BRD4) Interacts with Diverse Kinase Inhibitors.
Acs Chem.Biol., 9, 2014
|
|
4O72
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with NU7441 | | Descriptor: | 1,2-ETHANEDIOL, 8-(dibenzo[b,d]thiophen-4-yl)-2-(morpholin-4-yl)-4H-chromen-4-one, Bromodomain-containing protein 4, ... | | Authors: | Zhu, J.-Y, Ember, S.W, Watts, C, Schonbrunn, E. | | Deposit date: | 2013-12-24 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Acetyl-lysine Binding Site of Bromodomain-Containing Protein 4 (BRD4) Interacts with Diverse Kinase Inhibitors.
Acs Chem.Biol., 9, 2014
|
|
5LG7
 
 | | Solution NMR structure of Tryptophan to Arginine mutant of Arkadia RING domain | | Descriptor: | E3 ubiquitin-protein ligase Arkadia, ZINC ION | | Authors: | Birkou, M, Chasapis, C.T, Loutsidou, A.K, Bentrop, D, Lelli, M, Herrmann, T, Episkopou, V, Spyroulias, G.A. | | Deposit date: | 2016-07-06 | | Release date: | 2017-06-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A Residue Specific Insight into the Arkadia E3 Ubiquitin Ligase Activity and Conformational Plasticity.
J. Mol. Biol., 429, 2017
|
|
4CGZ
 
 | | Crystal structure of the Bloom's syndrome helicase BLM in complex with DNA | | Descriptor: | 5'-D(*AP*GP*CP*GP*TP*CP*GP*AP*GP*AP*TP*CP)-3', 5'-D(*GP*AP*TP*CP*TP*CP*GP*AP*CP*GP*CP*TP*CP*DT *CP*CP*CP)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Newman, J.A, Savitsky, P, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2013-11-27 | | Release date: | 2013-12-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of the Bloom'S Syndrome Helicase Indicates a Role for the Hrdc Domain in Conformational Changes.
Nucleic Acids Res., 43, 2015
|
|
4MEN
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a 5-methyl-triazolopyrimidine ligand | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, N,5-dimethyl-N-(4-methylbenzyl)[1,2,4]triazolo[1,5-a]pyrimidin-7-amine | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Martin, S, Fedorov, O, Vidler, L.R, Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Hoelder, S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-08-27 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Discovery of Novel Small-Molecule Inhibitors of BRD4 Using Structure-Based Virtual Screening.
J.Med.Chem., 56, 2013
|
|
4MI5
 
 | | Crystal structure of the EZH2 SET domain | | Descriptor: | Histone-lysine N-methyltransferase EZH2, SULFATE ION, ZINC ION | | Authors: | Antonysamy, S, Condon, B, Druzina, Z, Bonanno, J, Gheyi, T, Macewan, I, Zhang, A, Ashok, S, Russell, M, Luz, J.G. | | Deposit date: | 2013-08-30 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Context of Disease-Associated Mutations and Putative Mechanism of Autoinhibition Revealed by X-Ray Crystallographic Analysis of the EZH2-SET Domain.
Plos One, 8, 2013
|
|
4CL9
 
 | | N-TERMINAL BROMODOMAIN OF HUMAN BRD4 WITH I-BET295 | | Descriptor: | 1,2-ETHANEDIOL, BROMODOMAIN-CONTAINING PROTEIN 4, propan-2-yl N-[(2S,4R)-1-ethanoyl-2-methyl-6-[4-[[8-(oxidanylamino)-8-oxidanylidene-octanoyl]amino]phenyl]-3,4-dihydro-2H-quinolin-4-yl]carbamate | | Authors: | Chung, C, Atkinson, S. | | Deposit date: | 2014-01-13 | | Release date: | 2014-12-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Structure Based Design of Dual Hdac/Bet Inhibitors as Novel Epigenetic Probes.
Medchemcomm, 5, 2014
|
|
5LT4
 
 | |
5LUD
 
 | | Structure of Cyclophilin A in complex with 2,3-Diaminopyridine | | Descriptor: | Peptidyl-prolyl cis-trans isomerase, pyridine-2,3-diamine | | Authors: | McNae, I.W, Nowicki, M.W, Blackburn, E.A, Wear, M.A, Walkinshaw, M.D. | | Deposit date: | 2016-09-08 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Thermo-kinetic analysis space expansion for cyclophilin-ligand interactions - identification of a new nonpeptide inhibitor using BiacoreTM T200.
FEBS Open Bio, 7, 2017
|
|
4CDG
 
 | | Crystal structure of the Bloom's syndrome helicase BLM in complex with Nanobody | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BLOOM SYNDROME PROTEIN, NANOBODY, ... | | Authors: | Newman, J.A, Savitsky, P, Allerston, C.K, Pike, A.C.W, Pardon, E, Steyaert, J, Arrowsmith, C.H, von Delft, F, Bountra, C, Edwards, A, Gileadi, O. | | Deposit date: | 2013-10-31 | | Release date: | 2013-11-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.794 Å) | | Cite: | Crystal Structure of the Bloom'S Syndrome Helicase Indicates a Role for the Hrdc Domain in Conformational Changes.
Nucleic Acids Res., 43, 2015
|
|
4C5I
 
 | | Crystal structure of MBTD1 YY1 complex | | Descriptor: | MBT DOMAIN-CONTAINING PROTEIN 1, TRANSCRIPTIONAL REPRESSOR PROTEIN YY1 | | Authors: | Alfieri, C, Glatt, S, Mueller, C.W. | | Deposit date: | 2013-09-12 | | Release date: | 2013-11-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.586 Å) | | Cite: | Structural Basis for Targeting the Chromatin Repressor Sfmbt to Polycomb Response Elements
Genes Dev., 27, 2013
|
|
4BYJ
 
 | | Aurora A kinase bound to a highly selective imidazopyridine inhibitor | | Descriptor: | (S)-N-(1-(6-chloro-2-(1,3-dimethyl-1H-pyrazol-4-yl)-3H-imidazo[4,5-b]pyridin-7-yl)pyrrolidin-3-yl)acetamide, AURORA KINASE A | | Authors: | Joshi, A, Kosmopoulou, M, Bayliss, R. | | Deposit date: | 2013-07-19 | | Release date: | 2013-11-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Aurora Isoform Selectivity: Design and Synthesis of Imidazo[4,5-B]Pyridine Derivatives as Highly Selective Inhibitors of Aurora-A Kinase in Cells.
J.Med.Chem., 56, 2013
|
|
4A0V
 
 | | model refined against the Symmetry-free cryo-EM map of TRiC-AMP-PNP | | Descriptor: | T-COMPLEX PROTEIN 1 SUBUNIT BETA | | Authors: | Cong, Y, Schroder, G.F, Meyer, A.S, Jakana, J, Ma, B, Dougherty, M.T, Schmid, M.F, Reissmann, S, Levitt, M, Ludtke, S.L, Frydman, J, Chiu, W. | | Deposit date: | 2011-09-13 | | Release date: | 2012-02-15 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (10.7 Å) | | Cite: | Symmetry-Free Cryo-Em Structures of the Chaperonin Tric Along its ATPase-Driven Conformational Cycle.
Embo J., 31, 2012
|
|
5LJ2
 
 | | N-TERMINAL BROMODOMAIN OF HUMAN BRD4 WITH 5-(5-aminopyridin-3-yl)-8-(((3R,4R)-3-((1,1-dioxidotetrahydro-2H-thiopyran-4-yl)methoxy)piperidin-4-yl)amino)-3-methyl-1,7-naphthyridin-2(1H)-one | | Descriptor: | 1,2-ETHANEDIOL, 5-(5-aminopyridin-3-yl)-8-(((3R,4R)-3-((1,1-dioxidotetrahydro-2H-thiopyran-4-yl)methoxy)piperidin-4-yl)amino)-3-methyl-1,7-naphthyridin-2(1H)-one, Bromodomain-containing protein 4 | | Authors: | Chung, C, Bamborough, P, Demont, E. | | Deposit date: | 2016-07-17 | | Release date: | 2016-08-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | A Chemical Probe for the ATAD2 Bromodomain.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
4CCG
 
 | | Structure of an E2-E3 complex | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AMMONIUM ION, CHLORIDE ION, ... | | Authors: | Hodson, C, Purkiss, A, Walden, H. | | Deposit date: | 2013-10-22 | | Release date: | 2014-01-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the Human Fancl Ring-Ube2T Complex Reveals Determinants of Cognate E3-E2 Selection.
Structure, 22, 2014
|
|
5LHI
 
 | | Structure of the KDM1A/CoREST complex with the inhibitor N-[3-(ethoxymethyl)-2-[[4-[[(3R)-pyrrolidin-3-yl]methoxy]phenoxy]methyl]phenyl]-4-methylthieno[3,2-b]pyrrole-5-carboxamide | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1, ... | | Authors: | Cecatiello, V, Pasqualato, S. | | Deposit date: | 2016-07-12 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Thieno[3,2-b]pyrrole-5-carboxamides as New Reversible Inhibitors of Histone Lysine Demethylase KDM1A/LSD1. Part 2: Structure-Based Drug Design and Structure-Activity Relationship.
J. Med. Chem., 60, 2017
|
|
4A0K
 
 | | STRUCTURE OF DDB1-DDB2-CUL4A-RBX1 BOUND TO A 12 BP ABASIC SITE CONTAINING DNA-DUPLEX | | Descriptor: | 12 BP DNA, 12 BP THF CONTAINING DNA, CULLIN-4A, ... | | Authors: | Fischer, E.S, Scrima, A, Gut, H, Thoma, N.H. | | Deposit date: | 2011-09-09 | | Release date: | 2011-12-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (5.93 Å) | | Cite: | The Molecular Basis of Crl4(Ddb2/Csa) Ubiquitin Ligase Architecture, Targeting, and Activation.
Cell(Cambridge,Mass.), 147, 2011
|
|
3ZZY
 
 | |
