6PXY
 
 | |
3EHZ
 
 | |
4DSO
 
 | | Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, BENZAMIDINE, GLYCEROL, ... | | Authors: | Oh, A, Maurer, T, Garrenton, L.S, Pitts, K, Anderson, D.J, Skelton, N.J, Fauber, B.P, Pan, B, Malek, S, Stokoe, D, Ludlam, M, Bowman, K.K, Wu, J, Giannetti, A.M, Starovasnik, M.A, Mellman, I, Jackson, P.K, Ruldolph, J, Fang, G, Wang, W. | | Deposit date: | 2012-02-19 | | Release date: | 2012-04-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4DSU
 
 | | Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity | | Descriptor: | BENZIMIDAZOLE, GTPase KRas, isoform 2B, ... | | Authors: | Oh, A, Maurer, T, Garrenton, L.S, Pitts, K, Anderson, D.J, Skelton, N.J, Fauber, B.P, Pan, B, Malek, S, Stokoe, D, Ludlam, M, Bowman, K.K, Wu, J, Giannetti, A.M, Starovasnik, M.A, Mellman, I, Jackson, P.K, Ruldolph, J, Fang, G, Wang, W. | | Deposit date: | 2012-02-19 | | Release date: | 2012-04-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7T79
 
 | |
6PY4
 
 | |
7T78
 
 | |
7ZIY
 
 | | X-ray structure of the haloalkane dehalogenase HaloTag7 bound to a pentyltrifluoromethanesulfonamide tetramethylrhodamine ligand (TMR-T5) | | Descriptor: | CALCIUM ION, Haloalkane dehalogenase, [9-[2-carboxy-5-[2-[2-[5-(trifluoromethylsulfonylamino)pentoxy]ethoxy]ethylcarbamoyl]phenyl]-6-(dimethylamino)xanthen-3-ylidene]-dimethyl-azanium | | Authors: | Tarnawski, M, Kompa, J, Johnsson, K, Hiblot, J. | | Deposit date: | 2022-04-08 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Exchangeable HaloTag Ligands for Super-Resolution Fluorescence Microscopy.
J.Am.Chem.Soc., 145, 2023
|
|
6V4S
 
 | |
6PY5
 
 | |
2BU8
 
 | | crystal structures of human pyruvate dehydrogenase kinase 2 containing physiological and synthetic ligands | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DICHLORO-ACETIC ACID, MAGNESIUM ION, ... | | Authors: | Knoechel, T.R, Tucker, A.D, Robinson, C.M, Phillips, C, Taylor, W, Bungay, P.J, Kasten, S.A, Roche, T.E, Brown, D.G. | | Deposit date: | 2005-06-08 | | Release date: | 2006-02-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Regulatory Roles of the N-Terminal Domain Based on Crystal Structures of Human Pyruvate Dehydrogenase Kinase 2 Containing Physiological and Synthetic Ligands.
Biochemistry, 45, 2006
|
|
2BU2
 
 | | crystal structures of human pyruvate dehydrogenase kinase 2 containing physiological and synthetic ligands | | Descriptor: | 4-({(2R,5S)-2,5-DIMETHYL-4-[(2R)-3,3,3-TRIFLUORO-2-HYDROXY-2-METHYLPROPANOYL]PIPERAZIN-1-YL}CARBONYL)BENZONITRILE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Knoechel, T.R, Tucker, A.D, Robinson, C.M, Phillips, C, Taylor, W, Bungay, P.J, Kasten, S.A, Roche, T.E, Brown, D.G. | | Deposit date: | 2005-06-08 | | Release date: | 2006-02-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Regulatory Roles of the N-Terminal Domain Based on Crystal Structures of Human Pyruvate Dehydrogenase Kinase 2 Containing Physiological and Synthetic Ligands.
Biochemistry, 45, 2006
|
|
7R70
 
 | | Crystal Structure of the UbArk2C fusion protein | | Descriptor: | GLYCEROL, Ubiquitin,E3 ubiquitin-protein ligase RNF165, ZINC ION | | Authors: | Paluda, A, Middleton, A.J, Mace, P.D, Day, C.L. | | Deposit date: | 2021-06-24 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Ubiquitin and a charged loop regulate the ubiquitin E3 ligase activity of Ark2C.
Nat Commun, 13, 2022
|
|
2BTZ
 
 | | crystal structures of human pyruvate dehydrogenase kinase 2 containing physiological and synthetic ligands | | Descriptor: | PYRUVATE DEHYDROGENASE KINASE ISOENZYME 2 | | Authors: | Knoechel, T.R, Tucker, A.D, Robinson, C.M, Phillips, C, Taylor, W, Bungay, P.J, Kasten, S.A, Roche, T.E, Brown, D.G. | | Deposit date: | 2005-06-08 | | Release date: | 2006-02-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Regulatory Roles of the N-Terminal Domain Based on Crystal Structures of Human Pyruvate Dehydrogenase Kinase 2 Containing Physiological and Synthetic Ligands.
Biochemistry, 45, 2006
|
|
2BU6
 
 | | crystal structures of human pyruvate dehydrogenase kinase 2 containing physiological and synthetic ligands | | Descriptor: | (N-{4-[(ETHYLANILINO)SULFONYL]-2-METHYLPHENYL}-3,3,3-TRIFLUORO-2-HYDROXY-2-METHYLPROPANAMIDE, PYRUVATE DEHYDROGENASE KINASE ISOENZYME 2 | | Authors: | Knoechel, T.R, Tucker, A.D, Robinson, C.M, Phillips, C, Taylor, W, Bungay, P.J, Kasten, S.A, Roche, T.E, Brown, D.G. | | Deposit date: | 2005-06-08 | | Release date: | 2006-02-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Regulatory Roles of the N-Terminal Domain Based on Crystal Structures of Human Pyruvate Dehydrogenase Kinase 2 Containing Physiological and Synthetic Ligands.
Biochemistry, 45, 2006
|
|
6PYI
 
 | |
6Q0F
 
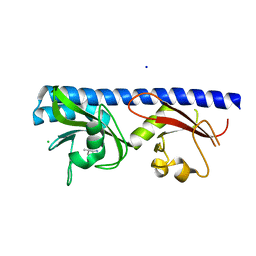 | | Crystal structure of ligand-binding domain of Pseudomonas fluorescens chemoreceptor CtaA in complex with L-valine | | Descriptor: | CHLORIDE ION, Putative methyl-accepting chemotaxis protein, SODIUM ION, ... | | Authors: | Ud-Din, I.A, Khan, M.F, Roujeinikova, A. | | Deposit date: | 2019-08-01 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Broad Specificity of Amino Acid Chemoreceptor CtaA ofPseudomonas fluorescensIs Afforded by Plasticity of Its Amphipathic Ligand-Binding Pocket.
Mol.Plant Microbe Interact., 33, 2020
|
|
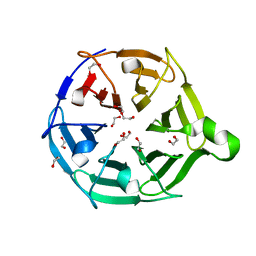
7QRX
 
 | |
2F00
 
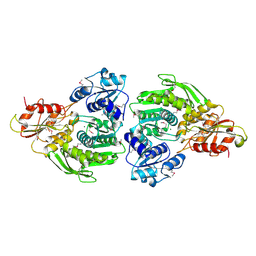 | | Escherichia coli MurC | | Descriptor: | MAGNESIUM ION, UDP-N-acetylmuramate--L-alanine ligase | | Authors: | Deva, T, Baker, E.N, Squire, C.J, Smith, C.A. | | Deposit date: | 2005-11-10 | | Release date: | 2006-10-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Escherichia coliUDP-N-acetylmuramoyl:L-alanine ligase (MurC).
Acta Crystallogr.,Sect.D, 62, 2006
|
|
1FBV
 
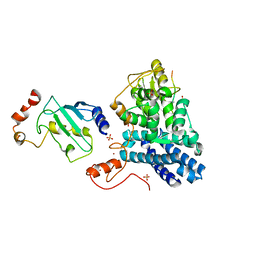 | | STRUCTURE OF A CBL-UBCH7 COMPLEX: RING DOMAIN FUNCTION IN UBIQUITIN-PROTEIN LIGASES | | Descriptor: | SIGNAL TRANSDUCTION PROTEIN CBL, SULFATE ION, UBIQUITIN-CONJUGATING ENZYME E12-18 KDA UBCH7, ... | | Authors: | Zheng, N, Wang, P, Jeffrey, P.D, Pavletich, N.P. | | Deposit date: | 2000-07-17 | | Release date: | 2000-08-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases.
Cell(Cambridge,Mass.), 102, 2000
|
|
2BU5
 
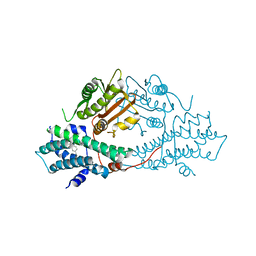 | | crystal structures of human pyruvate dehydrogenase kinase 2 containing physiological and synthetic ligands | | Descriptor: | 4-({(2R,5S)-2,5-DIMETHYL-4-[(2R)-3,3,3-TRIFLUORO-2-HYDROXY-2-METHYLPROPANOYL]PIPERAZIN-1-YL}CARBONYL)BENZONITRILE, PYRUVATE DEHYDROGENASE KINASE ISOENZYME 2 | | Authors: | Knoechel, T.R, Tucker, A.D, Robinson, C.M, Phillips, C, Taylor, W, Bungay, P.J, Kasten, S.A, Roche, T.E, Brown, D.G. | | Deposit date: | 2005-06-08 | | Release date: | 2006-02-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Regulatory Roles of the N-Terminal Domain Based on Crystal Structures of Human Pyruvate Dehydrogenase Kinase 2 Containing Physiological and Synthetic Ligands.
Biochemistry, 45, 2006
|
|
5HEO
 
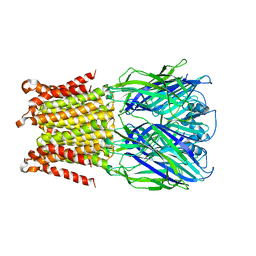 | |
5HPS
 
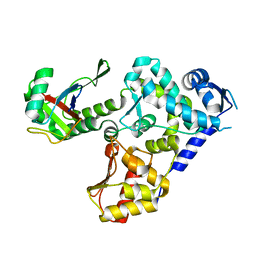 | |
6SWY
 
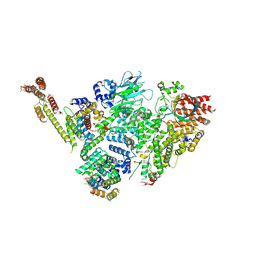 | | Structure of active GID E3 ubiquitin ligase complex minus Gid2 and delta Gid9 RING domain | | Descriptor: | Glucose-induced degradation protein 8, Protein FYV10,Protein FYV10,Protein FYV10,Protein FYV10,Protein FYV10,Protein FYV10,Protein FYV10, Vacuolar import and degradation protein 24, ... | | Authors: | Qiao, S, Prabu, J.R, Schulman, B.A. | | Deposit date: | 2019-09-24 | | Release date: | 2019-11-20 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Interconversion between Anticipatory and Active GID E3 Ubiquitin Ligase Conformations via Metabolically Driven Substrate Receptor Assembly
Mol.Cell, 77, 2020
|
|
4DST
 
 | | Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity | | Descriptor: | 1,2-ETHANEDIOL, 2-(4,6-dichloro-2-methyl-1H-indol-3-yl)ethanamine, ACETATE ION, ... | | Authors: | Oh, A, Maurer, T, Garrenton, L.S, Pitts, K, Anderson, D.J, Skelton, N.J, Fauber, B.P, Pan, B, Malek, S, Stokoe, D, Ludlam, M, Bowman, K.K, Wu, J, Giannetti, A.M, Starovasnik, M.A, Mellman, I, Jackson, P.K, Ruldolph, J, Fang, G, Wang, W. | | Deposit date: | 2012-02-19 | | Release date: | 2012-04-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
