1ROP
 
 | |
2JPX
 
 | | A18H Vpu TM structure in lipid bilayers | | Descriptor: | Vpu protein | | Authors: | Park, S, Opella, S.J. | | Deposit date: | 2007-05-25 | | Release date: | 2007-11-13 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Conformational changes induced by a single amino acid substitution in the trans-membrane domain of Vpu: implications for HIV-1 susceptibility to channel blocking drugs
Protein Sci., 16, 2007
|
|
4KSG
 
 | | Dna gyrase atp binding domain of enterococcus faecalis in complex with a small molecule inhibitor (4-[(1S,5R,6R)-6-AMINO-1-METHYL-3-AZABICYCLO[3.2.0]HEPT-3-YL]-6-FLUORO-N-METHYL-2-[(2-METHYLPYRIMIDIN-5-YL)OXY]-9H-PYRIMIDO[4,5-B]INDOL-8-AMINE) | | Descriptor: | 4-[(1S,5R,6R)-6-amino-1-methyl-3-azabicyclo[3.2.0]hept-3-yl]-6-fluoro-N-methyl-2-[(2-methylpyrimidin-5-yl)oxy]-9H-pyrimido[4,5-b]indol-8-amine, DNA gyrase subunit B, TERTIARY-BUTYL ALCOHOL | | Authors: | Bensen, D.C, Akers-Rodriguez, S, Tari, L.W. | | Deposit date: | 2013-05-17 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A new class of type iia topoisomerase inhibitors with
broad-spectrum antibacterial activity
To be Published
|
|
1E6V
 
 | | Methyl-coenzyme M reductase from Methanopyrus kandleri | | Descriptor: | 1-THIOETHANESULFONIC ACID, Coenzyme B, FACTOR 430, ... | | Authors: | Grabarse, W, Ermler, U. | | Deposit date: | 2000-08-23 | | Release date: | 2000-10-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Comparison of Three Methyl-Coenzyme M Reductases from Phylogenetically Distant Organisms: Unusual Amino Acid Modification, Conservation and Adaptation
J.Mol.Biol., 303, 2000
|
|
3VB3
 
 | | Crystal structure of SARS-CoV 3C-like protease in apo form | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, DI(HYDROXYETHYL)ETHER | | Authors: | Chuck, C.P, Wong, K.B. | | Deposit date: | 2011-12-31 | | Release date: | 2012-12-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design, synthesis and crystallographic analysis of nitrile-based broad-spectrum peptidomimetic inhibitors for coronavirus 3C-like proteases
Eur.J.Med.Chem., 59C, 2012
|
|
1J53
 
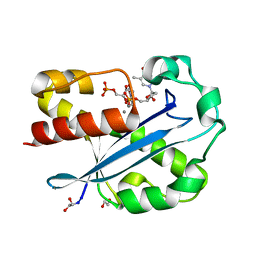 | | Structure of the N-terminal Exonuclease Domain of the Epsilon Subunit of E.coli DNA Polymerase III at pH 8.5 | | Descriptor: | 1,2-ETHANEDIOL, DNA polymerase III, epsilon chain, ... | | Authors: | Hamdan, S, Carr, P.D, Brown, S.E, Ollis, D.L, Dixon, N.E. | | Deposit date: | 2002-01-22 | | Release date: | 2002-10-16 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Proofreading during Replication of the Escherichia coli Chromosome
Structure, 10, 2002
|
|
282D
 
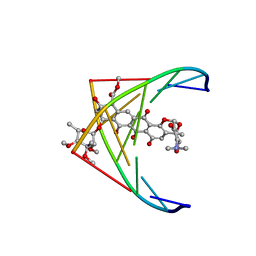 | | A CONTINOUS TRANSITION FROM A-DNA TO B-DNA IN THE 1:1 COMPLEX BETWEEN NOGALAMYCIN AND THE HEXAMER DCCCGGG | | Descriptor: | DNA (5'-D(*CP*CP*CP*GP*GP*G)-3'), NOGALAMYCIN | | Authors: | Cruse, W, Saludjian, P, Leroux, Y, Leger, Y, El Manouni, D, Prange, T. | | Deposit date: | 1996-08-26 | | Release date: | 1996-10-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A continuous transition from A-DNA to B-DNA in the 1:1 complex between nogalamycin and the hexamer dCCCGGG.
J.Biol.Chem., 271, 1996
|
|
3AT2
 
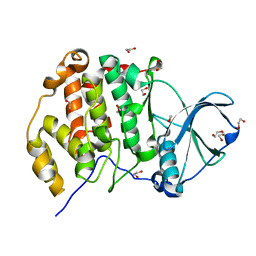 | | Crystal structure of CK2alpha | | Descriptor: | 1,2-ETHANEDIOL, Casein kinase II subunit alpha | | Authors: | Kinoshita, T. | | Deposit date: | 2010-12-23 | | Release date: | 2011-11-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A detailed thermodynamic profile of cyclopentyl and isopropyl derivatives binding to CK2 kinase
Mol.Cell.Biochem., 356, 2011
|
|
2DK7
 
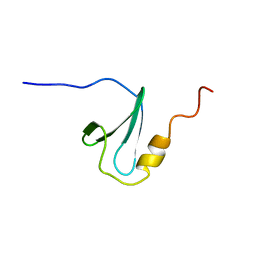 | | Solution structure of WW domain in transcription elongation regulator 1 | | Descriptor: | Transcription elongation regulator 1 | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-06 | | Release date: | 2007-04-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of WW domain in transcription elongation regulator 1
To be Published
|
|
4KTD
 
 | | Fab fragment of HIV vaccine-elicited CD4bs-directed antibody, GE136, from non-human primate | | Descriptor: | GE136 Heavy Chain Fab, GE136 Light Chain Fab, GLYCEROL, ... | | Authors: | Poulsen, C, Tran, K, Standfield, R, Wyatt, R.T. | | Deposit date: | 2013-05-20 | | Release date: | 2014-02-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Vaccine-elicited primate antibodies use a distinct approach to the HIV-1 primary receptor binding site informing vaccine redesign.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1IWD
 
 | |
1IYL
 
 | | Crystal Structure of Candida albicans N-myristoyltransferase with Non-peptidic Inhibitor | | Descriptor: | (1-METHYL-1H-IMIDAZOL-2-YL)-(3-METHYL-4-{3-[(PYRIDIN-3-YLMETHYL)-AMINO]-PROPOXY}-BENZOFURAN-2-YL)-METHANONE, Myristoyl-CoA:Protein N-Myristoyltransferase | | Authors: | Sogabe, S, Fukami, T.A, Morikami, K, Shiratori, Y, Aoki, Y, D'Arcy, A, Winkler, F.K, Banner, D.W, Ohtsuka, T. | | Deposit date: | 2002-08-29 | | Release date: | 2002-12-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structures of Candida albicans N-Myristoyltransferase with Two Distinct Inhibitors
CHEM.BIOL., 9, 2002
|
|
1EKV
 
 | | HUMAN BRANCHED CHAIN AMINO ACID AMINOTRANSFERASE (MITOCHONDRIAL): THREE DIMENSIONAL STRUCTURE OF ENZYME INACTIVATED BY TRIS BOUND TO THE PYRIDOXAL-5'-PHOSPHATE ON ONE END AND ACTIVE SITE LYS202 NZ ON THE OTHER. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BRANCHED CHAIN AMINO ACID AMINOTRANSFERASE (MITOCHONDRIAL), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Yennawar, N.H, Dunbar, J.H, Conway, M, Hutson, S.M, Farber, G.K. | | Deposit date: | 2000-03-09 | | Release date: | 2001-03-09 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The structure of human mitochondrial branched-chain aminotransferase.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1JLS
 
 | | STRUCTURE OF THE URACIL PHOSPHORIBOSYLTRANSFERASE URACIL/CPR 2 MUTANT C128V | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Schumacher, M.A, Bashor, C.J, Otsu, K, Zu, S, Parry, R, Ullman, B, Brennan, R.G. | | Deposit date: | 2001-07-16 | | Release date: | 2002-01-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structural mechanism of GTP stabilized oligomerization and catalytic activation of the Toxoplasma gondii uracil phosphoribosyltransferase.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
2QNL
 
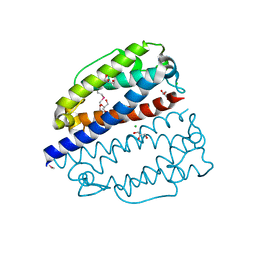 | |
1JOC
 
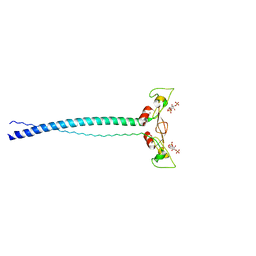 | | EEA1 homodimer of C-terminal FYVE domain bound to inositol 1,3-diphosphate | | Descriptor: | Early Endosomal Autoantigen 1, PHOSPHORIC ACID MONO-(2,3,4,6-TETRAHYDROXY-5-PHOSPHONOOXY-CYCLOHEXYL) ESTER, ZINC ION | | Authors: | Dumas, J.J, Merithew, E, Rajamani, D, Hayes, S, Lawe, D, Corvera, S, Lambright, D.G. | | Deposit date: | 2001-07-27 | | Release date: | 2001-12-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Multivalent endosome targeting by homodimeric EEA1.
Mol.Cell, 8, 2001
|
|
2G5X
 
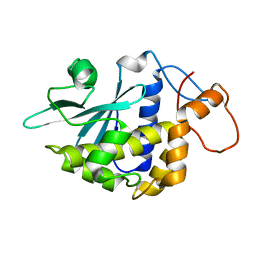 | | Crystal structure of lychnin a type 1 Ribosome Inactivating Protein (RIP) | | Descriptor: | Ribosome-inactivating protein | | Authors: | Fermani, S, Falini, G, Tosi, G, Ripamonti, A, Polito, L, Bolognesi, A, Stirpe, F. | | Deposit date: | 2006-02-23 | | Release date: | 2007-03-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of lychnin a type 1 Ribosome Inactivating Protein (RIP)
To be Published
|
|
1EIC
 
 | |
6JT4
 
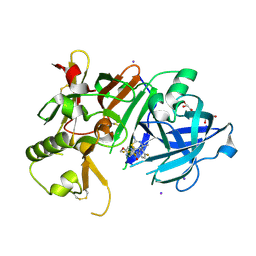 | | Crystal Structure of BACE1 in complex with N-{3-[(4S,6S)-2-amino-4-methyl-6-(trifluoromethyl)-5,6-dihydro-4H-1,3-thiazin-4-yl]-4-fluorophenyl}-5-(fluoromethoxy)pyrazine-2-carboxamide | | Descriptor: | Beta-secretase 1, GLYCEROL, IODIDE ION, ... | | Authors: | Anan, K, Iso, Y, Oguma, T, Nakahara, K, Suzuki, S, Yamamoto, T, Matsuoka, E, Ito, H, Sakaguchi, G, Ando, S, Morimoto, K, Kanegawa, N, Kido, Y, Kawachi, T, Fukushima, T, Teisman, A, Urmaliya, V, Dhuyvetter, D, Borghys, H, Austin, N, Bergh, A.V.D, Verboven, P, Bischoff, F, Gijsen, H.J.M, Yamano, Y, Kusakabe, K.I. | | Deposit date: | 2019-04-08 | | Release date: | 2020-01-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Trifluoromethyl Dihydrothiazine-Based beta-Secretase (BACE1) Inhibitors with Robust Central beta-Amyloid Reduction and Minimal Covalent Binding Burden.
Chemmedchem, 14, 2019
|
|
4LNB
 
 | | Aspergillus fumigatus protein farnesyltransferase ternary complex with farnesyldiphosphate and ethylenediamine scaffold inhibitor 5 | | Descriptor: | 1,2-ETHANEDIOL, CaaX farnesyltransferase alpha subunit Ram2, CaaX farnesyltransferase beta subunit Ram1, ... | | Authors: | Mabanglo, M.F, Hast, M.A, Beese, L.S. | | Deposit date: | 2013-07-11 | | Release date: | 2014-01-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Crystal structures of the fungal pathogen Aspergillus fumigatus protein farnesyltransferase complexed with substrates and inhibitors reveal features for antifungal drug design.
Protein Sci., 23, 2014
|
|
6JAL
 
 | | Crystal structure of ABC transporter alpha-glycoside-binding mutant protein R356A in ligand free form | | Descriptor: | 1,2-ETHANEDIOL, ABC transporter, periplasmic substrate-binding protein, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Gogoi, P. | | Deposit date: | 2019-01-24 | | Release date: | 2019-10-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural and thermodynamic correlation illuminates the selective transport mechanism of disaccharide alpha-glycosides through ABC transporter.
Febs J., 287, 2020
|
|
6CXS
 
 | | Crystal Structure of Clostridium perfringens beta-glucuronidase bound with a novel, potent inhibitor 4-(8-(piperazin-1-yl)-1,2,3,4-tetrahydro-[1,2,3]triazino[4',5':4,5]thieno[2,3-c]isoquinolin-5-yl)morpholine | | Descriptor: | 4-(8-(piperazin-1-yl)-1,2,3,4-tetrahydro-[1,2,3]triazino[4',5':4,5]thieno[2,3-c]isoquinolin-5-yl)morpholine, Beta-glucuronidase, Maltose/maltodextrin-binding periplasmic protein | | Authors: | Wallace, B.D, Redinbo, M.R. | | Deposit date: | 2018-04-04 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Targeted inhibition of gut bacterial beta-glucuronidase activity enhances anticancer drug efficacy.
Proc.Natl.Acad.Sci.USA, 2020
|
|
6D04
 
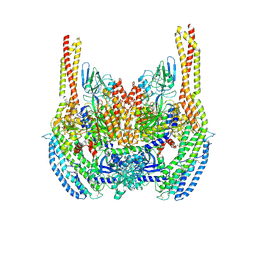 | | Cryo-EM structure of a Plasmodium vivax invasion complex essential for entry into human reticulocytes; two molecules of parasite ligand, subclass 1. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Gruszczyk, J, Huang, R.K, Hong, C, Yu, Z, Tham, W.H. | | Deposit date: | 2018-04-10 | | Release date: | 2018-06-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Cryo-EM structure of an essential Plasmodium vivax invasion complex.
Nature, 559, 2018
|
|
6JED
 
 | | Crystal structure of IMP-1 metallo-beta-lactamase in a complex with MCR | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Metallo-beta-lactamase type 2, SULFANYLACETIC ACID, ... | | Authors: | Wachino, J. | | Deposit date: | 2019-02-05 | | Release date: | 2019-08-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | 4-Amino-2-Sulfanylbenzoic Acid as a Potent Subclass B3 Metallo-beta-Lactamase-Specific Inhibitor Applicable for Distinguishing Metallo-beta-Lactamase Subclasses.
Antimicrob.Agents Chemother., 63, 2019
|
|
4E14
 
 | | Crystal structure of kynurenine formamidase conjugated with phenylmethylsulfonyl fluoride | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, kynurenine formamidase | | Authors: | Han, Q, Robinson, H, Li, J. | | Deposit date: | 2012-03-05 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Biochemical identification and crystal structure of kynurenine formamidase from Drosophila melanogaster.
Biochem.J., 446, 2012
|
|
