5OBB
 
 | |
5OKN
 
 | | Crystal structure of human SHIP2 Phosphatase-C2 D607A mutant | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, ... | | Authors: | Le Coq, J, Lietha, D. | | Deposit date: | 2017-07-25 | | Release date: | 2017-08-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis for interdomain communication in SHIP2 providing high phosphatase activity.
Elife, 6, 2017
|
|
7AVH
 
 | | Streptococcal High Identity Repeats in Tandem (SHIRT) domains 3-4 from cell surface protein SGO_0707 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, LPXTG cell wall surface protein | | Authors: | Whelan, F, Jenkins, H.T, Potts, J.R. | | Deposit date: | 2020-11-05 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Periscope Proteins are variable-length regulators of bacterial cell surface interactions.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5OLU
 
 | | The crystal structure of a highly thermostable carboxyl esterase from Bacillus coagulans in complex with glycerol | | Descriptor: | ACETATE ION, Alpha/beta hydrolase family protein, CHLORIDE ION, ... | | Authors: | Gourlay, L.J, Nakhnoukh, C, Bolognesi, M. | | Deposit date: | 2017-07-28 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A stereospecific carboxyl esterase from Bacillus coagulans hosting nonlipase activity within a lipase-like fold.
FEBS J., 285, 2018
|
|
5OCK
 
 | | Crystal structure of ACPA E4 in complex with CEP1 | | Descriptor: | CEP1 peptide (from enolase), Human ACPA E4 Fab fragment - Heavy chain, Human ACPA E4 Fab fragment - Light chain | | Authors: | Dobritzsch, D, Ge, C, Holmdahl, R. | | Deposit date: | 2017-07-03 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis of Cross-Reactivity of Anti-Citrullinated Protein Antibodies.
Arthritis Rheumatol, 71, 2019
|
|
5O0C
 
 | | Crystal structure of Phosphopantetheine adenylyltransferase from Mycobacterium abcessus in complex with 3-(3-methyl-1H-indol-1-yl)propanoic acid (Fragment 3) | | Descriptor: | 3-(3-methylindol-1-yl)propanoic acid, Phosphopantetheine adenylyltransferase | | Authors: | Thomas, S.E, Kim, S.Y, Mendes, V, Blaszczyk, M, Blundell, T.L. | | Deposit date: | 2017-05-16 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.642 Å) | | Cite: | Structural Biology and the Design of New Therapeutics: From HIV and Cancer to Mycobacterial Infections: A Paper Dedicated to John Kendrew.
J. Mol. Biol., 429, 2017
|
|
7AEI
 
 | | Studies Towards a Reversible EGFR C797S Triple Mutant Inhibitor Series | | Descriptor: | 5-chloranyl-~{N}2-[4-[4-(dimethylamino)piperidin-1-yl]-2-methoxy-5-(1-methylpyrazol-4-yl)phenyl]-~{N}4-(2-dimethylphosphorylphenyl)pyrimidine-2,4-diamine, Epidermal growth factor receptor | | Authors: | Hargreaves, D. | | Deposit date: | 2020-09-17 | | Release date: | 2021-06-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Abstract 4451: Evaluation of the therapeutic potential of phosphine oxide pyrazole inhibitors in tumors harboring EGFR C797S mutation
Cancer Res., 79, 2019
|
|
6UO6
 
 | |
6TVF
 
 | | Crystal structure of the haemagglutinin from a H10N7 seal influenza virus isolated in Germany in complex with human receptor analogue, 6'-SLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Zhang, J, Xiong, X, Purkiss, A, Walker, P, Gamblin, S, Skehel, J.J. | | Deposit date: | 2020-01-09 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Hemagglutinin Traits Determine Transmission of Avian A/H10N7 Influenza Virus between Mammals.
Cell Host Microbe, 28, 2020
|
|
6U99
 
 | | Hsp90a NTD covalently bound to sulfonyl fluoride probe 1 at K58 | | Descriptor: | 3-{[(3-{6-amino-8-[(6-iodo-2H-1,3-benzodioxol-5-yl)sulfanyl]-9H-purin-9-yl}propyl)amino]methyl}benzene-1-sulfinic acid, Heat shock protein HSP 90-alpha | | Authors: | Cuesta, A, Wan, X, Taunton, J. | | Deposit date: | 2019-09-06 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Ligand Conformational Bias Drives Enantioselective Modification of a Surface-Exposed Lysine on Hsp90.
J.Am.Chem.Soc., 142, 2020
|
|
6TVX
 
 | | Human CD73 (ecto 5'-nucleotidase) in complex with PSB12676 (an AOPCP derivative, compound 9 in paper) in the closed state | | Descriptor: | 5'-nucleotidase, ZINC ION, [[(2~{R},3~{S},4~{R},5~{R})-5-[2,6-bis(azanyl)purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]methylphosphonic acid | | Authors: | Pippel, J, Strater, N. | | Deposit date: | 2020-01-10 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 2-Substituted alpha , beta-Methylene-ADP Derivatives: Potent Competitive Ecto-5'-nucleotidase (CD73) Inhibitors with Variable Binding Modes.
J.Med.Chem., 63, 2020
|
|
6TWH
 
 | | Crystal structure of the haemagglutinin mutant (Gln226Leu, Gly228Ser) from an H10N7 seal influenza virus isolated in Germany | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Zhang, J, Xiong, X, Purkiss, A, Walker, P, Gamblin, S, Skehel, J.J. | | Deposit date: | 2020-01-13 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Hemagglutinin Traits Determine Transmission of Avian A/H10N7 Influenza Virus between Mammals.
Cell Host Microbe, 28, 2020
|
|
5OE7
 
 | |
5OEB
 
 | | Structure of large terminase from the thermophilic bacteriophage D6E in complex with ADP (Crystal form 3) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Large subunit terminase, MAGNESIUM ION | | Authors: | Xu, R.G, Jenkins, H.T, Greive, S.J, Antson, A.A. | | Deposit date: | 2017-07-07 | | Release date: | 2017-10-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the large terminase from a hyperthermophilic virus reveals a unique mechanism for oligomerization and ATP hydrolysis.
Nucleic Acids Res., 45, 2017
|
|
6TX5
 
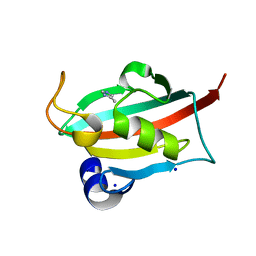 | |
7AIZ
 
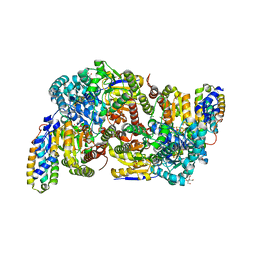 | | Vanadium nitrogenase VFe protein, high CO state | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-HYDROXY-3-CARBOXY-ADIPIC ACID, ... | | Authors: | Rohde, M, Einsle, O. | | Deposit date: | 2020-09-28 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Two ligand-binding sites in CO-reducing V nitrogenase reveal a general mechanistic principle.
Sci Adv, 7, 2021
|
|
7AYF
 
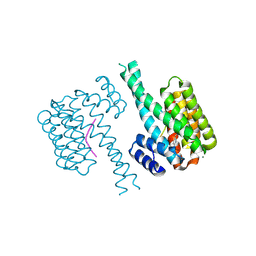 | | 14-3-3 sigma with Pin1 binding site pS72 and covalently bound TCF521-110 | | Descriptor: | 14-3-3 protein sigma, 6-(2-bromanylimidazol-1-yl)pyridine-3-carbaldehyde, CALCIUM ION, ... | | Authors: | Wolter, M, Dijck, L.v, Ottmann, C. | | Deposit date: | 2020-11-12 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
6UBZ
 
 | | Crystal structure of D678A GoxA bound to glycine at pH 5.5 | | Descriptor: | GLYCINE, MAGNESIUM ION, Uncharacterized protein GoxA | | Authors: | Yukl, E.T. | | Deposit date: | 2019-09-13 | | Release date: | 2019-10-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Kinetic and structural evidence that Asp-678 plays multiple roles in catalysis by the quinoprotein glycine oxidase.
J.Biol.Chem., 294, 2019
|
|
7AXN
 
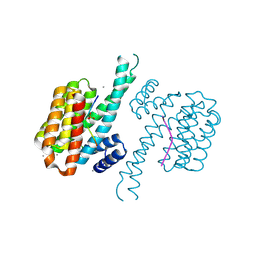 | | 14-3-3 sigma in complex with Pin1 binding site pS72 and covalently bound TCF521-026 | | Descriptor: | 14-3-3 protein sigma, 3-chloranyl-4-imidazol-1-yl-benzaldehyde, CALCIUM ION, ... | | Authors: | Wolter, M, Dijck, L.v, Ottmann, C. | | Deposit date: | 2020-11-10 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
5O32
 
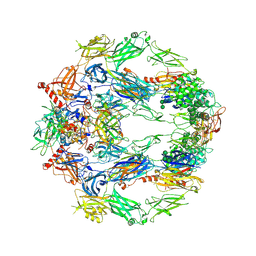 | | The structure of complement complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Xue, X, Wu, J, Forneris, F, Gros, P. | | Deposit date: | 2017-05-23 | | Release date: | 2017-06-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (4.206209 Å) | | Cite: | Regulator-dependent mechanisms of C3b processing by factor I allow differentiation of immune responses.
Nat. Struct. Mol. Biol., 24, 2017
|
|
7AKR
 
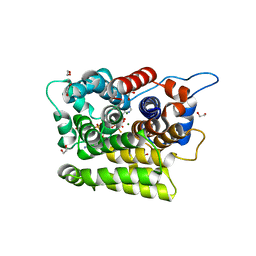 | |
5OG5
 
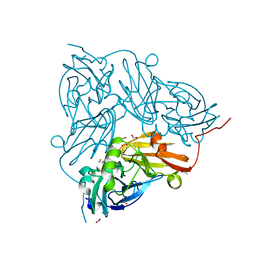 | | Cu nitrite reductase serial data at varying temperatures RT 0.21MGy | | Descriptor: | ACETATE ION, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Horrell, S, Kekilli, D, Strange, R.W, Hough, M.A. | | Deposit date: | 2017-07-11 | | Release date: | 2018-05-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Enzyme catalysis captured using multiple structures from one crystal at varying temperatures.
IUCrJ, 5, 2018
|
|
5O4T
 
 | |
7AZW
 
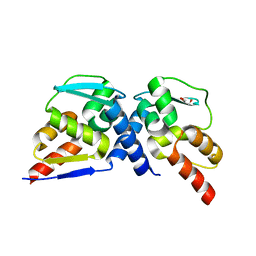 | | Crystal structure of the MIZ1-BTB-domain | | Descriptor: | GLYCEROL, Zinc finger and BTB domain-containing protein 17 isoform X1 | | Authors: | Orth, B, Sander, B, Diederichs, K, Lorenz, S. | | Deposit date: | 2020-11-17 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of an atypical interaction site in the BTB domain of the MYC-interacting zinc-finger protein 1.
Structure, 29, 2021
|
|
7AZ2
 
 | | 14-3-3 sigma with Pin1 binding site pS72 and covalently bound LvD1014 | | Descriptor: | 1-[4-methyl-2-(trifluoromethyl)phenyl]-2-phenyl-imidazole, 14-3-3 protein sigma, CALCIUM ION, ... | | Authors: | Wolter, M, Dijck, L.v, Ottmann, C. | | Deposit date: | 2020-11-14 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.081 Å) | | Cite: | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
