6FV1
 
 | | Structure of human coronavirus NL63 main protease in complex with the alpha-ketoamide (S)-N-((S)-4-(benzylamino)-3,4-dioxo-1-((S)-2-oxopyrrolidin-3-yl)butan-2-yl)-2-cinnamamido-4-methylpentanamide (cinnamoyl-leucine-GlnLactam-CO-CO-NH-benzyl) | | Descriptor: | (2~{S})-4-methyl-~{N}-[(2~{S},3~{R})-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]-2-[[(~{E})-3-phenylprop-2-enoyl]amino]pentanamide, 3C-like proteinase, DIMETHYL SULFOXIDE, ... | | Authors: | Zhang, L, Hilgenfeld, R. | | Deposit date: | 2018-02-28 | | Release date: | 2019-03-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Alpha-ketoamides as broad-spectrum inhibitors of coronavirus and enterovirus replication Structure-based design, synthesis, and activity assessment.
J.Med.Chem., 2020
|
|
4Y5F
 
 | | PAS-GAF fragment from Deinococcus radiodurans BphP assembled with BV - Y307S, high dose | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome | | Authors: | Li, F, Burgie, E.S, Yu, T, Heroux, A, Schatz, G.C, Vierstra, R.D, Orville, A.M. | | Deposit date: | 2015-02-11 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray radiation induces deprotonation of the bilin chromophore in crystalline D. radiodurans phytochrome.
J.Am.Chem.Soc., 137, 2015
|
|
1Q05
 
 | | Crystal structure of the Cu(I) form of E. coli CueR, a copper efflux regulator | | Descriptor: | COPPER (I) ION, Transcriptional regulator cueR | | Authors: | Changela, A, Chen, K, Xue, Y, Holschen, J, Outten, C.E, O'Halloran, T.V, Mondragon, A. | | Deposit date: | 2003-07-15 | | Release date: | 2003-09-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR
Science, 301, 2003
|
|
8IZE
 
 | | Crystal structure of intracellular B30.2 domain of BTN3A1 in complex with 4-HMBPP | | Descriptor: | Butyrophilin subfamily 3 member A1, DI(HYDROXYETHYL)ETHER, [(E)-3-(hydroxymethyl)pent-2-enyl] phosphono hydrogen phosphate | | Authors: | Yang, Y.Y, Yi, S.M, Huang, J.W, Chen, C.C, Guo, R.T. | | Deposit date: | 2023-04-07 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Phosphoantigens glue butyrophilin 3A1 and 2A1 to activate V gamma 9V delta 2 T cells.
Nature, 621, 2023
|
|
2X90
 
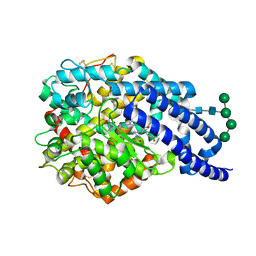 | | Crystal structure of AnCE-enalaprilat complex | | Descriptor: | 1-((2S)-2-{[(1S)-1-CARBOXY-3-PHENYLPROPYL]AMINO}PROPANOYL)-L-PROLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Akif, M, Georgiadis, D, Mahajan, A, Dive, V, Sturrock, E.D, Isaac, R.E, Acharya, K.R. | | Deposit date: | 2010-03-14 | | Release date: | 2010-06-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | High Resolution Crystal Structures of Drosophila Melanogaster Angiotensin Converting Enzyme in Complex with Novel Inhibitors and Anti- Hypertensive Drugs.
J.Mol.Biol., 400, 2010
|
|
2X8Z
 
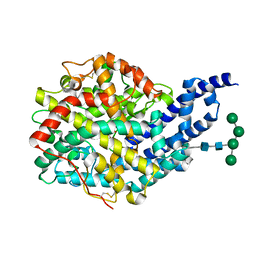 | | Crystal structure of AnCE-captopril complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ANGIOTENSIN CONVERTING ENZYME, L-CAPTOPRIL, ... | | Authors: | Akif, M, Georgiadis, D, Mahajan, A, Dive, V, Sturrock, E.D, Isaac, R.E, Acharya, K.R. | | Deposit date: | 2010-03-14 | | Release date: | 2010-06-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | High Resolution Crystal Structures of Drosophila Melanogaster Angiotensin Converting Enzyme in Complex with Novel Inhibitors and Anti- Hypertensive Drugs.
J.Mol.Biol., 400, 2010
|
|
4IAQ
 
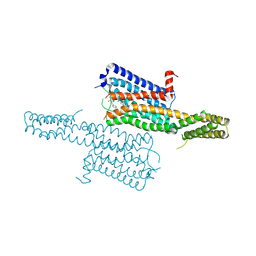 | | Crystal structure of the chimeric protein of 5-HT1B-BRIL in complex with dihydroergotamine (PSI Community Target) | | Descriptor: | Chimera protein of human 5-hydroxytryptamine receptor 1B and E. Coli soluble cytochrome b562, Dihydroergotamine | | Authors: | Wang, C, Jiang, Y, Ma, J, Wu, H, Wacker, D, Katritch, V, Han, G.W, Liu, W, Huang, X, Vardy, E, McCorvy, J.D, Gao, X, Zhou, E.X, Melcher, K, Zhang, C, Bai, F, Yang, H, Yang, L, Jiang, H, Roth, B.L, Cherezov, V, Stevens, R.C, Xu, H.E, GPCR Network (GPCR) | | Deposit date: | 2012-12-07 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for molecular recognition at serotonin receptors.
Science, 340, 2013
|
|
5EJE
 
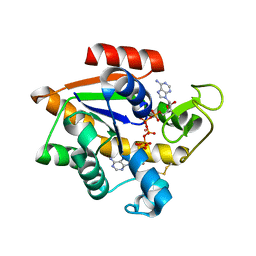 | | Crystal structure of E. coli Adenylate kinase G56C/T163C double mutant in complex with Ap5a | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, COBALT (II) ION | | Authors: | Sauer, U.H, Kovermann, M, Grundstrom, C, Wolf-Watz, M, Sauer-Eriksson, A.E. | | Deposit date: | 2015-11-01 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for ligand binding to an enzyme by a conformational selection pathway.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6G7M
 
 | | Four-site variant (Y222C, C197S, C432S, C433S) of E. coli hydrogenase-2 | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, Hydrogenase-2 large chain, ... | | Authors: | Carr, S.B, Armstrong, F.A, Zhang, L, Beaton, S.E. | | Deposit date: | 2018-04-06 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Direct visible light activation of a surface cysteine-engineered [NiFe]-hydrogenase by silver nanoclusters
Energy Environ Sci, 2019
|
|
1ISZ
 
 | | Crystal structure of xylanase from Streptomyces olivaceoviridis E-86 complexed with galactose | | Descriptor: | beta-D-galactopyranose, endo-1,4-beta-D-xylanase | | Authors: | Fujimoto, Z, Kuno, A, Kaneko, S, Kobayashi, H, Kusakabe, I, Mizuno, H. | | Deposit date: | 2001-12-27 | | Release date: | 2002-02-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the sugar complexes of Streptomyces olivaceoviridis E-86 xylanase: sugar binding structure of the family 13 carbohydrate binding module.
J.Mol.Biol., 316, 2002
|
|
5O4X
 
 | | Protein structure determination by electron diffraction using a single three-dimensional nanocrystal | | Descriptor: | Lysozyme C | | Authors: | Clabbers, M.T.B, van Genderen, E, Wan, W, Wiegers, E.L, Gruene, T, Abrahams, J.P. | | Deposit date: | 2017-05-31 | | Release date: | 2017-08-23 | | Last modified: | 2024-01-17 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.11 Å) | | Cite: | Protein structure determination by electron diffraction using a single three-dimensional nanocrystal.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
1GEU
 
 | | ANATOMY OF AN ENGINEERED NAD-BINDING SITE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLUTATHIONE REDUCTASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Mittl, P.R.E, Schulz, G.E. | | Deposit date: | 1994-01-18 | | Release date: | 1994-11-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Anatomy of an engineered NAD-binding site.
Protein Sci., 3, 1994
|
|
5N5O
 
 | |
3UUS
 
 | |
1JDU
 
 | | CRYSTAL STRUCTURE OF 5'-DEOXY-5'-METHYLTHIOADENOSINE PHOSPHORYLASE | | Descriptor: | 5'-METHYLTHIOADENOSINE PHOSPHORYLASE | | Authors: | Appleby, T.C, Mathews, I.I, Porcelli, M, Cacciapuoti, G, Ealick, S.E. | | Deposit date: | 2001-06-15 | | Release date: | 2001-10-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-dimensional structure of a hyperthermophilic 5'-deoxy-5'-methylthioadenosine phosphorylase from Sulfolobus solfataricus.
J.Biol.Chem., 276, 2001
|
|
1JAW
 
 | | AMINOPEPTIDASE P FROM E. COLI LOW PH FORM | | Descriptor: | ACETATE ION, AMINOPEPTIDASE P, MANGANESE (II) ION | | Authors: | Wilce, M.C.J, Bond, C.S, Lilley, P.E, Dixon, N.E, Freeman, H.C, Guss, J.M. | | Deposit date: | 1997-12-22 | | Release date: | 1999-04-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and mechanism of a proline-specific aminopeptidase from Escherichia coli.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1GET
 
 | | ANATOMY OF AN ENGINEERED NAD-BINDING SITE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLUTATHIONE REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Mittl, P.R.E, Schulz, G.E. | | Deposit date: | 1994-01-18 | | Release date: | 1994-11-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Anatomy of an engineered NAD-binding site.
Protein Sci., 3, 1994
|
|
1GFZ
 
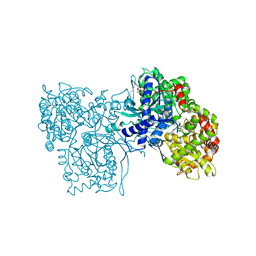 | | FLAVOPIRIDOL INHIBITS GLYCOGEN PHOSPHORYLASE BY BINDING AT THE INHIBITOR SITE | | Descriptor: | CAFFEINE, GLYCOGEN PHOSPHORYLASE, INOSINIC ACID, ... | | Authors: | Oikonomakos, N.G, Zographos, S.E, Skamnaki, V.T, Tsitsanou, K.E, Johnson, L.N. | | Deposit date: | 2000-06-29 | | Release date: | 2000-07-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Flavopiridol inhibits glycogen phosphorylase by binding at the inhibitor site.
J.Biol.Chem., 275, 2000
|
|
2VVQ
 
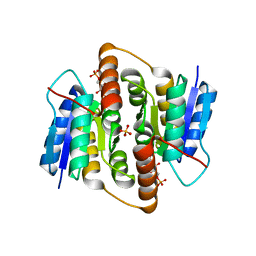 | | Crystal structure of Mycobacterium tuberculosis ribose-5-phosphate isomerase B in complex with the inhibitor 5-deoxy-5-phospho-D- ribonate | | Descriptor: | 5-O-phosphono-D-ribonic acid, RIBOSE-5-PHOSPHATE ISOMERASE B, SULFATE ION | | Authors: | Kowalinski, E, Roos, A.K, Mariano, S, Salmon, L, Mowbray, S.L. | | Deposit date: | 2008-06-10 | | Release date: | 2008-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | D-Ribose-5-Phosphate Isomerase B from Escherichia Coli is Also a Functional D-Allose-6-Phosphate Isomerase, While the Mycobacterium Tuberculosis Enzyme is not.
J.Mol.Biol., 382, 2008
|
|
1GES
 
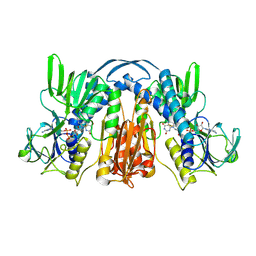 | |
5YXC
 
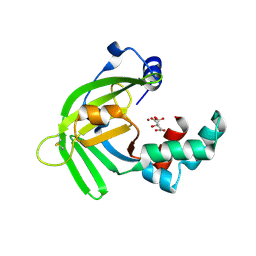 | | Crystal structure of Zinc binding protein ZinT in complex with citrate from E. coli | | Descriptor: | CITRIC ACID, Metal-binding protein ZinT, ZINC ION | | Authors: | Chen, J, Wang, L, Shang, F, Xu, Y. | | Deposit date: | 2017-12-04 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.763 Å) | | Cite: | Crystal structure of E. coli ZinT with one zinc-binding mode and complexed with citrate
Biochem. Biophys. Res. Commun., 500, 2018
|
|
6GMO
 
 | |
4N56
 
 | |
4N5S
 
 | |
1I7X
 
 | | BETA-CATENIN/E-CADHERIN COMPLEX | | Descriptor: | BETA-CATENIN, EPITHELIAL-CADHERIN | | Authors: | Huber, A.H, Weis, W.I. | | Deposit date: | 2001-03-10 | | Release date: | 2001-05-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of the beta-catenin/E-cadherin complex and the molecular basis of diverse ligand recognition by beta-catenin.
Cell(Cambridge,Mass.), 105, 2001
|
|
