4H2P
 
 | | Tetrameric form of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase (MHPCO) | | Descriptor: | 1,2-ETHANEDIOL, 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, BETA-MERCAPTOETHANOL, ... | | Authors: | Kobayashi, J, Yoshida, H, Mikami, B, Hayashi, H, Kamitori, S, Yagi, T. | | Deposit date: | 2012-09-13 | | Release date: | 2013-09-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | Crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase
To be Published
|
|
5EME
 
 | | Complex of RNA r(GCAGCAGC) with antisense PNA p(CTGCTGC) | | Descriptor: | Antisense PNA strand, CHLORIDE ION, RNA (5'-R(*GP*CP*AP*GP*CP*AP*GP*C)-3') | | Authors: | Kiliszek, A, Banaszak, K, Dauter, Z, Rypniewski, W. | | Deposit date: | 2015-11-06 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | The first crystal structures of RNA-PNA duplexes and a PNA-PNA duplex containing mismatches-toward anti-sense therapy against TREDs.
Nucleic Acids Res., 44, 2016
|
|
8WHU
 
 | | Spike Trimer of BA.2.86 in complex with two hACE2s | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Processed angiotensin-converting enzyme 2, ... | | Authors: | Yue, C, Liu, P. | | Deposit date: | 2023-09-23 | | Release date: | 2024-07-03 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
2BR4
 
 | | cmcI-D160 Mg-SAM | | Descriptor: | CEPHALOSPORIN HYDROXYLASE CMCI, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Oster, L.M, Lester, D.R, Terwisscha van Scheltinga, A, Svenda, M, Genereux, C, Andersson, I. | | Deposit date: | 2005-05-01 | | Release date: | 2006-03-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Insights Into Cephamycin Biosynthesis: The Crystal Structure of Cmci from Streptomyces Clavuligerus.
J.Mol.Biol., 358, 2006
|
|
5EN8
 
 | |
4H3Y
 
 | |
4KVK
 
 | | Crystal structure of Oryza sativa fatty acid alpha-dioxygenase | | Descriptor: | CALCIUM ION, CHLORIDE ION, DECYL-BETA-D-MALTOPYRANOSIDE, ... | | Authors: | Zhu, G, Koszelak-Rosenblum, M, Malkowski, M.G. | | Deposit date: | 2013-05-22 | | Release date: | 2013-08-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structures of alpha-dioxygenase from Oryza sativa: Insights into substrate binding and activation by hydrogen peroxide.
Protein Sci., 22, 2013
|
|
5EOM
 
 | | Structure of full-length human MAB21L1 with bound CTP | | Descriptor: | CITRIC ACID, CYTIDINE-5'-TRIPHOSPHATE, Protein mab-21-like 1, ... | | Authors: | de Oliveira Mann, C.C, Witte, G, Hopfner, K.-P. | | Deposit date: | 2015-11-10 | | Release date: | 2016-06-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural and biochemical characterization of the cell fate determining nucleotidyltransferase fold protein MAB21L1.
Sci Rep, 6, 2016
|
|
3DV0
 
 | | Snapshots of catalysis in the E1 subunit of the pyruvate dehydrogenase multi-enzyme complex | | Descriptor: | 2-{4-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-3-METHYLTHIOPHEN-2-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, MAGNESIUM ION, ... | | Authors: | Pei, X.Y, Titman, C.M, Frank, R.A.W, Leeper, F.J, Luisi, B.F. | | Deposit date: | 2008-07-18 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Snapshots of catalysis in the e1 subunit of the pyruvate dehydrogenase multienzyme complex
Structure, 16, 2008
|
|
4GWA
 
 | | Crystal Structure of a GH7 Family Cellobiohydrolase from Limnoria quadripunctata | | Descriptor: | GH7 family protein, MAGNESIUM ION | | Authors: | McGeehan, J.E, Martin, R.N.A, Streeter, S.D, Cragg, S.M, Guille, M.J, Schnorr, K.M, Kern, M, Bruce, N.C, McQueen-Mason, S.J. | | Deposit date: | 2012-09-01 | | Release date: | 2013-06-12 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural characterization of a unique marine animal family 7 cellobiohydrolase suggests a mechanism of cellulase salt tolerance
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4KZF
 
 | | The mechanism of the amidases: The effect of the mutation E142L in the amidase from Geobacillus pallidus | | Descriptor: | Aliphatic amidase, CHLORIDE ION | | Authors: | Weber, B.W, Sewell, B.T, Kimani, S.W, Varsani, A, Cowan, D.A, Hunter, R. | | Deposit date: | 2013-05-29 | | Release date: | 2013-08-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The mechanism of the amidases: mutating the glutamate adjacent to the catalytic triad inactivates the enzyme due to substrate mispositioning.
J.Biol.Chem., 288, 2013
|
|
4KWG
 
 | | Crystal Structure Analysis of ALDH2+ALDiB13 | | Descriptor: | 1,2-ETHANEDIOL, 7-bromo-5-methyl-1H-indole-2,3-dione, Aldehyde dehydrogenase, ... | | Authors: | Hurley, T.D, Kimble-Hill, A.C. | | Deposit date: | 2013-05-24 | | Release date: | 2014-04-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Development of selective inhibitors for aldehyde dehydrogenases based on substituted indole-2,3-diones.
J.Med.Chem., 57, 2014
|
|
5EPT
 
 | |
4KZK
 
 | |
8WXF
 
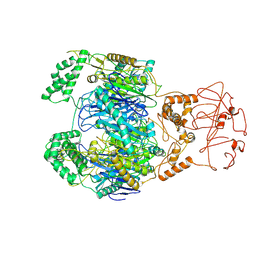 | | PNPase of Mycobacterium tuberculosis | | Descriptor: | Bifunctional guanosine pentaphosphate synthetase/polyribonucleotide nucleotidyltransferase | | Authors: | Wang, N, Sheng, Y.N, Liu, Y.T. | | Deposit date: | 2023-10-29 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures of Mycobacterium tuberculosis polynucleotide phosphorylase suggest a potential mechanism for its RNA substrate degradation.
Arch.Biochem.Biophys., 754, 2024
|
|
2VQT
 
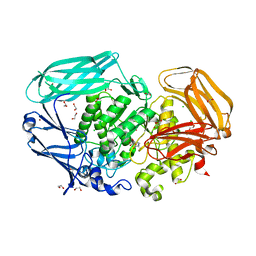 | | Structural and biochemical evidence for a boat-like transition state in beta-mannosidases | | Descriptor: | (2Z,3R,4S,5R,6R)-2-[(2-aminoethyl)imino]-6-(hydroxymethyl)piperidine-3,4,5-triol, 1,2-ETHANEDIOL, BETA-MANNOSIDASE, ... | | Authors: | Tailford, L.E, Offen, W.A, Smith, N.L, Dumon, C, Moreland, C, Gratien, J, Heck, M.P, Stick, R.V, Bleriot, Y, Vasella, A, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2008-03-18 | | Release date: | 2008-04-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Biochemical Evidence for a Boat-Like Transition State in Beta-Mannosidases.
Nat.Chem.Biol., 4, 2008
|
|
7L6N
 
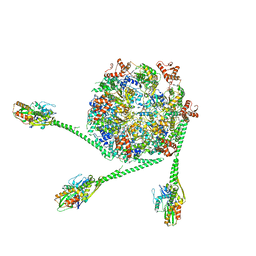 | | The Mycobacterium tuberculosis ClpB disaggregase hexamer structure with three locally refined ClpB middle domains and three DnaK nucleotide binding domains | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperone protein ClpB, Chaperone protein DnaK, ... | | Authors: | Yin, Y.Y, Feng, X, Li, H. | | Deposit date: | 2020-12-23 | | Release date: | 2021-05-26 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Structural basis for aggregate dissolution and refolding by the Mycobacterium tuberculosis ClpB-DnaK bi-chaperone system.
Cell Rep, 35, 2021
|
|
7L0Z
 
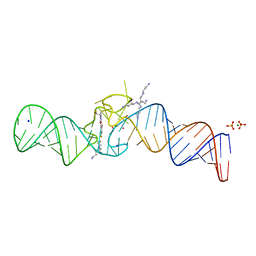 | | Spinach variant bound to DFHBI-1T | | Descriptor: | (5Z)-5-(3,5-difluoro-4-hydroxybenzylidene)-2-methyl-3-(2,2,2-trifluoroethyl)-3,5-dihydro-4H-imidazol-4-one, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Truong, L, Trachman, R.J, Ferre-D'Amare, A.R. | | Deposit date: | 2020-12-13 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Fluorogenic aptamers resolve the flexibility of RNA junctions using orientation-dependent FRET.
Rna, 27, 2021
|
|
3DZC
 
 | | 2.35 Angstrom resolution structure of WecB (VC0917), a UDP-N-acetylglucosamine 2-epimerase from Vibrio cholerae. | | Descriptor: | CALCIUM ION, CHLORIDE ION, UDP-N-acetylglucosamine 2-epimerase | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Kwon, K, Hasseman, J, Peterson, S.N, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-07-29 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | 2.35 Angstrom resolution structure of WecB (VC0917), a UDP-N-acetylglucosamine 2-epimerase from Vibrio cholerae.
TO BE PUBLISHED
|
|
5EDT
 
 | | Crystal structure of Mycobacterium tuberculosis CYP121 in complex with LIG9 | | Descriptor: | 2-azanyl-5-chloranyl-benzamide, Cytochrome P450 121, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Kavanagh, M.E, Coyne, A.G, McLean, K.J, James, G.G, Levy, C, Marino, L.B, Carvalho, L.P.D, Chan, D.S.H, Hudson, S.A, Surade, S, Munro, A.W, Abell, C. | | Deposit date: | 2015-10-22 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Fragment-Based Approaches to the Development of Mycobacterium tuberculosis CYP121 Inhibitors.
J.Med.Chem., 59, 2016
|
|
8WWP
 
 | | PNPase mutant of Mycobacterium tuberculosis | | Descriptor: | Bifunctional guanosine pentaphosphate synthetase/polyribonucleotide nucleotidyltransferase | | Authors: | Wang, N, Sheng, Y.N, Liu, Y.T. | | Deposit date: | 2023-10-26 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Cryo-EM structures of Mycobacterium tuberculosis polynucleotide phosphorylase suggest a potential mechanism for its RNA substrate degradation.
Arch.Biochem.Biophys., 754, 2024
|
|
8VFF
 
 | | Binary DNA Polymerase Beta bound to DNA containing primer terminal FapydG base-paired with a dA | | Descriptor: | DNA (5'-D(*CP*CP*GP*AP*CP*GP*AP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*(FAP))-3'), DNA (5'-D(P*GP*TP*CP*GP*G)-3'), ... | | Authors: | Oden, P.N, Ryan, B.J, Freudenthal, B.D. | | Deposit date: | 2023-12-21 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Biochemical and structural characterization of Fapy•dG replication by Human DNA polymerase beta.
Nucleic Acids Res., 52, 2024
|
|
3DZE
 
 | | Crystal structure of bovine coupling Factor B bound with cadmium | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ATP synthase subunit s, mitochondrial, ... | | Authors: | Lee, J.K, Stroud, R.M, Belogrudov, G.I. | | Deposit date: | 2008-07-29 | | Release date: | 2008-08-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal structure of bovine mitochondrial factor B at 0.96-A resolution.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
5EEV
 
 | | RADIATION DAMAGE TO THE TRAP-RNA COMPLEX: DOSE (DWD) 3.88 MGy | | Descriptor: | (GAGUU)10GAG 53-NUCLEOTIDE RNA, TRYPTOPHAN, Transcription attenuation protein MtrB | | Authors: | Bury, C.S, McGeehan, J.E, Garman, E.F, Shevtsov, M.B. | | Deposit date: | 2015-10-23 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | RNA protects a nucleoprotein complex against radiation damage.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
4GYL
 
 | | The E142L mutant of the amidase from Geobacillus pallidus showing the result of Michael addition of acrylamide at the active site cysteine | | Descriptor: | Aliphatic amidase, CHLORIDE ION, PROPIONAMIDE | | Authors: | Weber, B.W, Sewell, B.T, Kimani, S.W, Varsani, A, Cowan, D.A, Hunter, R. | | Deposit date: | 2012-09-05 | | Release date: | 2013-08-21 | | Last modified: | 2014-02-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The mechanism of the amidases: mutating the glutamate adjacent to the catalytic triad inactivates the enzyme due to substrate mispositioning.
J.Biol.Chem., 288, 2013
|
|
