8JWV
 
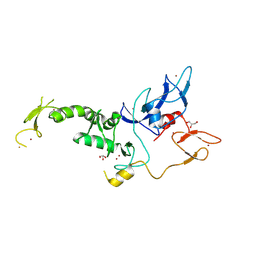 | | Untethered R0RBR | | Descriptor: | BARIUM ION, E3 ubiquitin-protein ligase parkin, GLYCEROL, ... | | Authors: | Lenka, D.R, Kumar, A. | | Deposit date: | 2023-06-29 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A new feedforward mechanism of Parkin activation.
To Be Published
|
|
8JWK
 
 | | The second purified state crystal structure of AKRtyl | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Aldo/keto reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Lin, S, Dai, S, Xiao, Z. | | Deposit date: | 2023-06-29 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | A three-level regulatory mechanism of the aldo-keto reductase subfamily AKR12D.
Nat Commun, 15, 2024
|
|
8JWJ
 
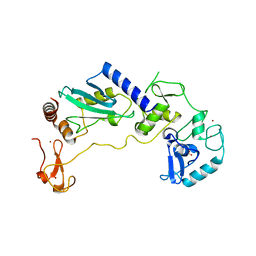 | | PHD Finger Protein 7 (PHF7) in complex with UBE2D2 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, PHD finger protein 7, ... | | Authors: | Lee, H.S, Bang, I, Choi, H.-J. | | Deposit date: | 2023-06-29 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Molecular basis for PHF7-mediated ubiquitination of histone H3.
Genes Dev., 37, 2023
|
|
8JWD
 
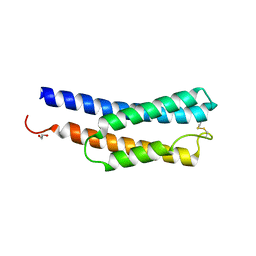 | | Histidine kinase QseE sensor domain of Escherichia coli O157:H7 | | Descriptor: | 1,2-ETHANEDIOL, histidine kinase | | Authors: | Matsumoto, K, Fukuda, Y, Inoue, T. | | Deposit date: | 2023-06-28 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Crystal structures of QseE and QseG: elements of a three-component system from Escherichia coli.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
8JW3
 
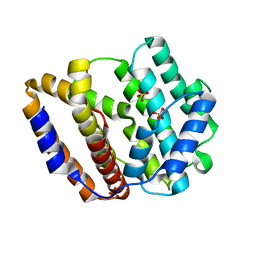 | | The crystal structure of the viral terpene synthase from Orpheovirus IHUMI-LCC2 | | Descriptor: | SULFATE ION, Terpenoid synthase | | Authors: | Jung, Y, Mitsuhashi, T, Senda, M, Sato, S, Senda, T, Fujita, M. | | Deposit date: | 2023-06-28 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Function and Structure of a Terpene Synthase Encoded in a Giant Virus Genome.
J.Am.Chem.Soc., 145, 2023
|
|
8JW0
 
 | | PSI-AcpPCI supercomplex from Amphidinium carterae | | Descriptor: | (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Li, Z.H, Li, X.Y, Wang, W.D. | | Deposit date: | 2023-06-28 | | Release date: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures and organizations of PSI-AcpPCI supercomplexes from red tidal and coral symbiotic photosynthetic dinoflagellates.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8JVZ
 
 | |
8JVY
 
 | |
8JVX
 
 | |
8JVW
 
 | |
8JVV
 
 | |
8JVU
 
 | |
8JVT
 
 | |
8JVS
 
 | |
8JVR
 
 | |
8JVQ
 
 | |
8JVP
 
 | |
8JVO
 
 | |
8JVN
 
 | |
8JVG
 
 | | Crystal structure of dephospho-coenzyme A kinase | | Descriptor: | GTP-dependent dephospho-CoA kinase | | Authors: | Kita, A, Ishida, Y, Shimosaka, T, Michimori, Y, Makarova, K, Koonin, E, Atomi, H, Miki, K. | | Deposit date: | 2023-06-28 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of GTP-dependent dephospho-coenzyme A kinase from the hyperthermophilic archaeon, Thermococcus kodakarensis.
Proteins, 92, 2024
|
|
8JVF
 
 | | Crystal structure of dephospho-coenzyme A kinase | | Descriptor: | GTP-dependent dephospho-CoA kinase, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Kita, A, Ishida, Y, Shimosaka, T, Michimori, Y, Makarova, K, Koonin, E, Atomi, H, Miki, K. | | Deposit date: | 2023-06-28 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of GTP-dependent dephospho-coenzyme A kinase from the hyperthermophilic archaeon, Thermococcus kodakarensis.
Proteins, 92, 2024
|
|
8JVC
 
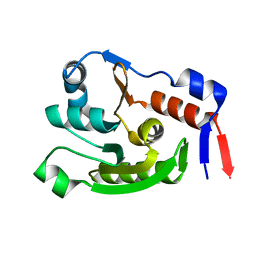 | | Crystal structure of dephospho-coenzyme A kinase | | Descriptor: | GTP-dependent dephospho-CoA kinase | | Authors: | Kita, A, Ishida, Y, Shimosaka, T, Michimori, Y, Makarova, K, Koonin, E, Atomi, H, Miki, K. | | Deposit date: | 2023-06-28 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of GTP-dependent dephospho-coenzyme A kinase from the hyperthermophilic archaeon, Thermococcus kodakarensis.
Proteins, 92, 2024
|
|
8JV8
 
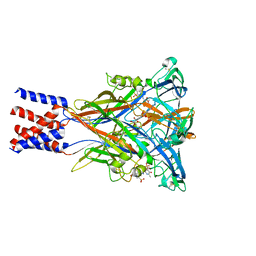 | | Cryo-EM structure of the panda P2X7 receptor in complex with PPNDS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[(E)-{4-formyl-5-hydroxy-6-methyl-3-[(phosphonooxy)methyl]pyridin-2-yl}diazenyl]-7-nitronaphthalene-1,5-disulfonic acid, P2X purinoceptor | | Authors: | Sheng, D, Hattori, M. | | Deposit date: | 2023-06-27 | | Release date: | 2023-11-29 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structural insights into the orthosteric inhibition of P2X receptors by non-ATP analog antagonists.
Elife, 12, 2024
|
|
8JV7
 
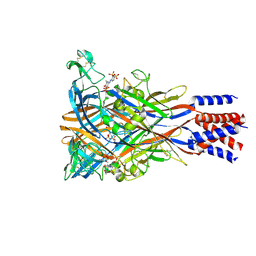 | | Cryo-EM structure of the panda P2X7 receptor in complex with PPADS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[(E)-[4-methanoyl-6-methyl-5-oxidanyl-3-(phosphonooxymethyl)pyridin-2-yl]diazenyl]benzene-1,3-disulfonic acid, P2X purinoceptor | | Authors: | Sheng, D, Hattori, M. | | Deposit date: | 2023-06-27 | | Release date: | 2023-11-29 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insights into the orthosteric inhibition of P2X receptors by non-ATP analog antagonists.
Elife, 12, 2024
|
|
8JV5
 
 | |
